
In molecular biology, Small nucleolar RNA R105/R108 refers to a group of related non-coding RNA (ncRNA) molecules which function in the biogenesis of other small nuclear RNAs (snRNAs). These small nucleolar RNAs (snoRNAs) are modifying RNAs and usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis.
In molecular biology, Small nucleolar RNA Z41 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA. snoRNA Z41 belongs to the C/D box class of snoRNAs which contain the conserved sequence motifs known as the C box (UGAUGA) and the D box (CUGA). Most of the members of the box C/D family function in directing site-specific 2'-O-methylation of substrate RNAs. Plant snoRNA Z41 was identified in screens of Arabidopsis thaliana.

In molecular biology, small nucleolar RNA R71 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA.
In molecular biology, R64/Z200 is a member of the C/D class of small nucleolar RNA which guide the site-specific 2'-O-methylation of substrate RNA. This family can be found in Arabidopsis thaliana (R64) and Oryza sativa (Z200).
In molecular biology, the snoRNA snoZ7/snoZ77 family contains related non-coding RNA molecules that are members of the C/D class of snoRNA which contain the C box motif (UGAUGA) and the D box motif (CUGA). Most of the members of the box C/D family function in directing site-specific 2'-O-methylation of substrate RNAs. snoZ7 RNA guides the methylation of 25S rRNA position A1372, whereas SnoR77Y guides the methylation of 18S rRNA at position U580. This family of snoRNAs has been found in plant species such as Arabidopsis thaliana.
In molecular biology, Small nucleolar RNA snR61/Z1/Z11 refers to a group of related non-coding RNA (ncRNA) molecules which function in the biogenesis of other small nuclear RNAs (snRNAs). These small nucleolar RNAs (snoRNAs) are modifying RNAs and usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis.
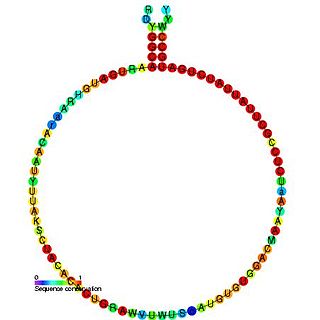
In molecular biology, Small nucleolar RNA R16 is a non-coding RNA (ncRNA) molecule identified in plants which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA.
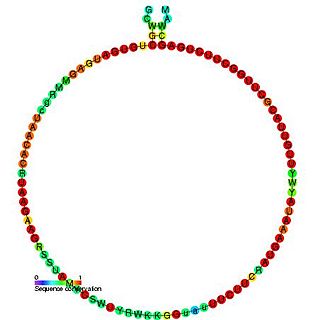
In molecular biology, Small nucleolar RNA R24 is a non-coding RNA (ncRNA) molecule identified in plants which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA.
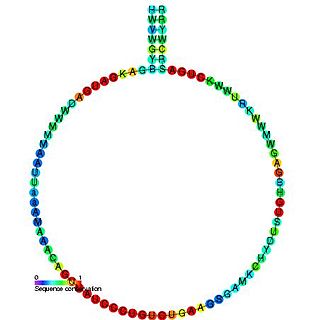
In molecular biology, Small nucleolar RNA R38 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA. snoRNA R38 belongs to the C/D box class of snoRNAs which contain the conserved sequence motifs known as the C box (UGAUGA) and the D box (CUGA). Most of the members of the box C/D family function in directing site-specific 2'-O-methylation of substrate RNAs. snoRNA R38 has been identified in human, yeast, Arabidopsis thaliana and Oryza sativa . snoRNA R38 guides the methylation of 2'-O-ribose sites in 28S rRNA.
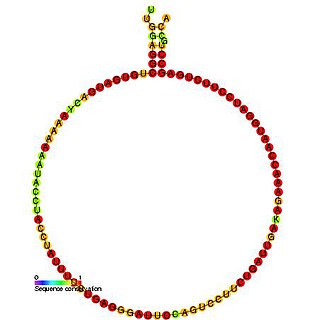
In molecular biology, Small nucleolar RNA R41 is a non-coding RNA (ncRNA) molecule identified in plants which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA.

In molecular biology, Small nucleolar RNA R79 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA. snoRNA R79 belongs to the C/D box class of snoRNAs which contain the conserved sequence motifs known as the C box (UGAUGA) and the D box (CUGA). Most of the members of the box C/D family function in directing site-specific 2'-O-methylation of substrate RNAs. Plant snoRNA R79 was identified in a screen of Arabidopsis thaliana.

In molecular biology, Small nucleolar RNA snoR86 is a non-coding RNA (ncRNA) which modifies other small nuclear RNAs (snRNAs). It is a member of the H/ACA class of small nucleolar RNA that guide the sites of modification of uridines to pseudouridines. Plant snoR86 was identified in a screen of Arabidopsis thaliana.
In molecular biology, Small nucleolar RNA snoR98 is a non-coding RNA (ncRNA) which modifies other small nuclear RNAs (snRNAs). It is a member of the H/ACA class of small nucleolar RNA that guide the sites of modification of uridines to pseudouridines. Plant snoR98 was identified in a screen of Arabidopsis thaliana.

In molecular biology, Small nucleolar RNA Z195/SNORD33 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA.
In molecular biology, Small nucleolar RNA Z196/R39/R59 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA.

In molecular biology, SNORD19 is a non-coding RNA (ncRNA) molecule which functions in the biogenesis (modification) of other small nuclear RNAs (snRNAs). This type of modifying RNA is located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA.

In molecular biology, snoRNA U36 is a non-coding RNA (ncRNA) molecule which functions in the biogenesis (modification) of other small nuclear RNAs (snRNAs). This type of modifying RNA is located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA.
In molecular biology, snoRNA U46 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA.

In molecular biology, snoRNA U59 is an RNA molecule that belongs to the C/D class of snoRNA, which contain the C box motif (UGAUGA) and the D box motif (CUGA). Most of the members of the box C/D family function in directing site-specific 2'-O-methylation of substrate RNAs. There are two closely related copies of U59, called SNORD59A and SNORD59B. They are both expressed from the intron of the host gene ATP5A. Both SNORD59A and SNORD59B target the 2'-O-methylation of 18S rRNA position A1031. This RNA has been identified in both the human and mouse genomes. Not to be confused with the plant snoRNA U59.
In molecular biology, Small nucleolar RNA Z223 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA.




















