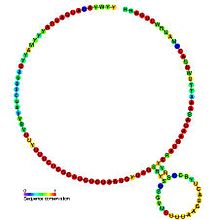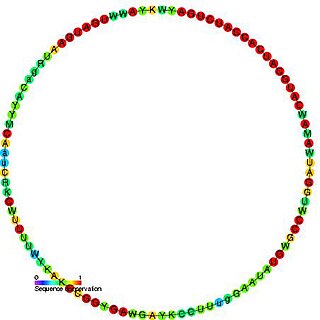
In molecular biology, Small nucleolar RNA snR55/Z10 is a non-coding RNA (ncRNA) molecule which functions in the biogenesis of other small nuclear RNAs (snRNAs). These small nucleolar RNAs (snoRNAs) are modifying RNAs and usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis.
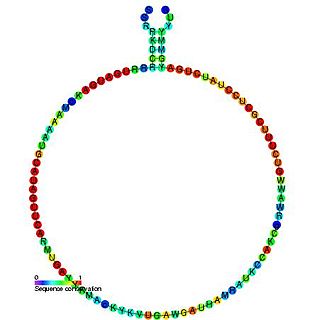
In molecular biology, snoRNA snR60 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA. snoRNA snR60 belongs to the C/D box class of snoRNAs which contain the conserved sequence motifs known as the C box (UGAUGA) and the D box (CUGA). Most of the members of the box C/D family function in directing site-specific 2'-O-methylation of substrate RNAs. snoRNA snR60 was initially discovered using a computational screen of the Saccharomyces cerevisiae genome, subsequent experimental screens discovered plant homologues Z15, Z230, Z193 and J17 and human U80/SNORD80.
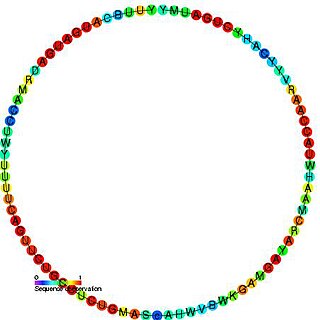
In molecular biology, Small nucleolar RNA snR61/Z1/Z11 refers to a group of related non-coding RNA (ncRNA) molecules which function in the biogenesis of other small nuclear RNAs (snRNAs). These small nucleolar RNAs (snoRNAs) are modifying RNAs and usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis.

In molecular biology, Small nucleolar RNA snR52 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA. snoRNA Z13 belongs to the C/D box class of snoRNAs which contain the conserved sequence motifs known as the C box (UGAUGA) and the D box (CUGA). Most of the members of the box C/D family function in directing site-specific 2'-O-methylation of substrate RNAs.
In molecular biology, Small nucleolar RNA Me28S-Gm1083 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA. snoRNA Me28S-Gm1083 belongs to the C/D box class of snoRNAs which contain the conserved sequence motifs known as the C box (UGAUGA) and the D box (CUGA). Most of the members of the box C/D family function in directing site-specific 2'-O-methylation of substrate RNAs. It is predicted that this family directs 2'-O-methylation of 28S G-1083.

In molecular biology, Small nucleolar RNA R12 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA.
In molecular biology, Small nucleolar RNA R160 is a non-coding RNA (ncRNA) molecule identified in plants which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA.
In molecular biology, Small nucleolar RNA R38 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA. snoRNA R38 belongs to the C/D box class of snoRNAs which contain the conserved sequence motifs known as the C box (UGAUGA) and the D box (CUGA). Most of the members of the box C/D family function in directing site-specific 2'-O-methylation of substrate RNAs. snoRNA R38 has been identified in human, yeast, Arabidopsis thaliana and Oryza sativa . snoRNA R38 guides the methylation of 2'-O-ribose sites in 28S rRNA.
In molecular biology, Small nucleolar RNA R41 is a non-coding RNA (ncRNA) molecule identified in plants which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA.

In molecular biology, Small nucleolar RNA R66 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA.
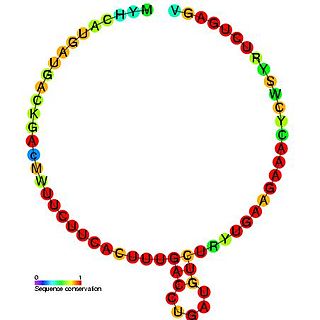
In molecular biology, SNORD37 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA.
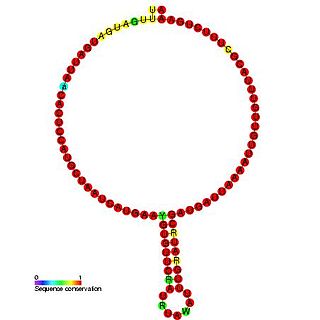
In molecular biology,snoRNA snR53 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA. snoRNA snR53 belongs to the C/D box class of snoRNAs which contain the conserved sequence motifs known as the C box (UGAUGA) and the D box (CUGA). Most of the members of the box C/D family function in directing site-specific 2'-O-methylation of substrate RNAs. snoRNA snR53 was initially discovered using a computational screen of the Saccharomyces cerevisiae genome.
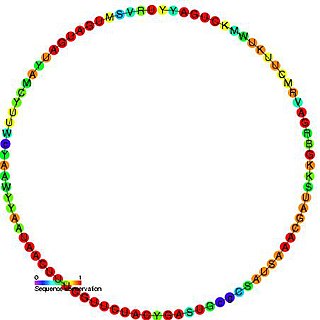
In molecular biology, snR54 is a non-coding RNA that is a member of the C/D class of snoRNA which contain the C box motif (UGAUGA) and D box motif (CUGA). Most of the members of the box C/D family function in directing site-specific 2'-O-methylation of substrate RNAs. This snoRNA was first identified by a computational screen followed by experimental verification. This RNA guides the 2'-O-methylation of 18S rRNA. In yeast this snoRNA is found to reside in an intron of the IMD4 gene.
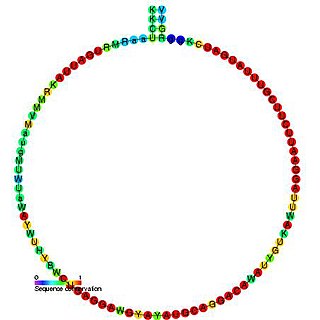
In molecular biology, snoRNA snR57 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA. snoRNA snR57 belongs to the C/D box class of snoRNAs which contain the conserved sequence motifs known as the C box (UGAUGA) and the D box (CUGA). Most of the members of the box C/D family function in directing site-specific 2'-O-methylation of substrate RNAs. snoRNA snR57 was initially discovered using a computational screen of the Saccharomyces cerevisiae genome.
In molecular biology, snR64 is an RNA molecule belonging to the C/D class of small nucleolar RNA (snoRNA), which contain the C (UGAUGA) and D (CUGA) box motifs. Similar to most members of the box C/D family, snR64 is conjectured to help direct site-specific 2'-O-methylation of substrate RNAs.
snoRNA snR66 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA. snoRNA snR66 belongs to the C/D box class of snoRNAs which contain the conserved sequence motifs known as the C box (UGAUGA) and the D box (CUGA). Most of the members of the box C/D family function in directing site-specific 2'-O-methylation of substrate RNAs. snoRNA snR66 was initially discovered using a computational screen of the Saccharomyces cerevisiae genome.
In molecular biology, snoRNA snR69 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA. snoRNA snR69 belongs to the C/D box class of snoRNAs which contain the conserved sequence motifs known as the C box (UGAUGA) and the D box (CUGA). Most of the members of the box C/D family function in directing site-specific 2'-O-methylation of substrate RNAs. snoRNA snR69 was initially discovered using a computational screen of the Saccharomyces cerevisiae genome.
In molecular biology, snoRNA snR71 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA. snoRNA snR71 belongs to the C/D box class of snoRNAs which contain the conserved sequence motifs known as the C box (UGAUGA) and the D box (CUGA). Most of the members of the box C/D family function in directing site-specific 2'-O-methylation of substrate RNAs. snoRNA snR71 was initially discovered using a computational screen of the Saccharomyces cerevisiae genome.
In molecular biology, snoR9 is a non-coding RNA (ncRNA) which functions in the biogenesis (modification) of other small nuclear RNAs (snRNAs). It is known as a small nucleolar RNA (snoRNA) and also often referred to as a 'guide RNA'.
The small nucleolar RNAs (snoRNAs) represent an abundant group of small non-coding RNAs (ncRNAs) in eukaryotes. With the exception of RNase MRP, all the snoRNAs fall into two major families, box C/D and box H/ACA snoRNAs, on the basis of common sequence motifs and structural features. They can be divided into guide and orphan snoRNAs according to the presence or absence of antisense sequence to rRNAs or snRNAs. Recently, thousands of novel snoRNAs were identified from small RNA sequencing datasets in human, mouse, chicken, Ciona intestinalis, Drosophila melanogaster, Caenhorhabditis elegans and Arabidopsis thaliana using snoSeekerNGS program.