
Prader–Willi syndrome (PWS) is a genetic disorder caused by a loss of function of specific genes on chromosome 15. In newborns, symptoms include weak muscles, poor feeding, and slow development. Beginning in childhood, those affected become constantly hungry, which often leads to obesity and type 2 diabetes. Mild to moderate intellectual impairment and behavioral problems are also typical of the disorder. Often, affected individuals have a narrow forehead, small hands and feet, short height, and light skin and hair. Most are unable to have children.
In molecular biology, Small nucleolar RNAs (snoRNAs) are a class of small RNA molecules that primarily guide chemical modifications of other RNAs, mainly ribosomal RNAs, transfer RNAs and small nuclear RNAs. There are two main classes of snoRNA, the C/D box snoRNAs, which are associated with methylation, and the H/ACA box snoRNAs, which are associated with pseudouridylation. SnoRNAs are commonly referred to as guide RNAs but should not be confused with the guide RNAs that direct RNA editing in trypanosomes or the guide RNAs (gRNAs) used by Cas9 for CRISPR gene editing.

In molecular biology, for Homo sapienssnoRA35 is an H/ACA box snoRNA, first cloned from a mouse adult brain cDNA library by Cavaillé et al. (2000), and found to be specifically expressed in the choroid plexus. Its human orthologue, HBI-36 was discovered by a homology search, and was found to be specifically expressed in the brain. Its gene resides in the second intron of the serotonin receptor 2c (5HT-2c) gene, which is predominantly expressed in choroid plexus epithelial cells. The human 5HT-2c mRNA was predicted to be 2'O-methylated by the C/D box snoRNP HBII-52 at a position also subjected to A:I editing. HBI-36 has no documented RNA target.

In molecular biology, SNORD116 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA.

In molecular biology, snoRNA U16 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA.

In molecular biology, SNORD19 is a non-coding RNA (ncRNA) molecule which functions in the biogenesis (modification) of other small nuclear RNAs (snRNAs). This type of modifying RNA is located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA.
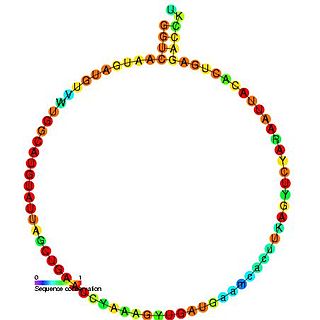
In molecular biology, snoRNA U45 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA.

In molecular biology, snoRNA U50 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA.

In molecular biology, snoRNA U59 is an RNA molecule that belongs to the C/D class of snoRNA, which contain the C box motif (UGAUGA) and the D box motif (CUGA). Most of the members of the box C/D family function in directing site-specific 2'-O-methylation of substrate RNAs. There are two closely related copies of U59, called SNORD59A and SNORD59B. They are both expressed from the intron of the host gene ATP5A. Both SNORD59A and SNORD59B target the 2'-O-methylation of 18S rRNA position A1031. This RNA has been identified in both the human and mouse genomes. Not to be confused with the plant snoRNA U59.

In molecular biology, SNORD64 is a non-coding RNA (ncRNA) molecule which functions in the biogenesis (modification) of other small nuclear RNAs (snRNAs). This type of modifying RNA is located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA.

In molecular biology, SNORD66 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA.

In molecular biology, snoRNA SNORD70 (HBII-234) is a non-coding RNA that belongs to the C/D family of snoRNAs. It is the human orthologue of the mouse MBII-234 and is predicted to guide 2'O-ribose methylation of the small 18S rRNA on position A512. It is hosted, together with HBII-95, by the core C/D box snoRNA protein encoding gene NOP5/NOP58.
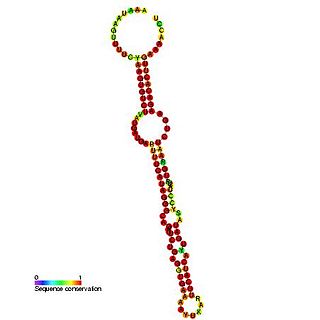
In molecular biology, snoRNA SNORD90 (HBII-295) is a non-coding RNA that belongs to the family of C/D snoRNAs. Initially described as HBII-295 this RNA has now been called SNORD70 by the HUGO Gene Nomenclature Committee. It is the human orthologue of the mouse MBII-295 and has no identified RNA target. This RNA is expressed from an intron of the MNAB/OR1K1 gene.

In molecular biology, Small Nucleolar RNA SNORD93 is a non-coding RNA (ncRNA) molecule that functions in the biogenesis (modification) of other small nuclear RNAs (snRNAs). This type of modifying RNA is located in the nucleolus of the Eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and is also often referred to as a guide RNA.

In molecular biology, Small Nucleolar RNA SNORD23 is a non-coding RNA (ncRNA) molecule which functions in the biogenesis (modification) of other small nuclear RNAs (snRNAs). This type of modifying RNA is located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA.

In molecular biology, Small Nucleolar RNA SNORD75 is a non-coding RNA (ncRNA) molecule which functions in the biogenesis (modification) of other small nuclear RNAs (snRNAs). This type of modifying RNA is located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA.

In molecular biology, Small Nucleolar RNA SNORD88 is a non-coding RNA (ncRNA) molecule which functions in the biogenesis (modification) of other small nuclear RNAs (snRNAs). This type of modifying RNA is located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA.

In molecular biology, Small Nucleolar RNA SNORD92 is a non-coding RNA (ncRNA) molecule which functions in the biogenesis (modification) of other small nuclear RNAs (snRNAs). This type of modifying RNA is located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA.
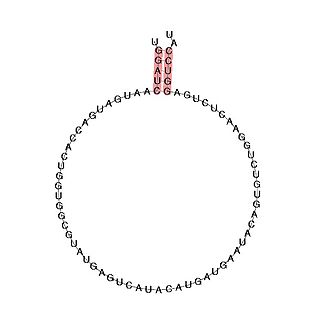
In molecular biology, Small nucleolar RNA SNORD113 is a small nucleolar RNA molecule which is located in the imprinted human 14q32 locus and may play a role in the evolution and/or mechanism of the epigenetic imprinting process.

UBE3A-ATS/Ube3a-ATS (human/mouse), otherwise known as ubiquitin ligase E3A-ATS, is the name for the antisense DNA strand that is transcribed as part of a larger transcript called LNCAT at the Ube3a locus. The Ube3a locus is imprinted and in the central nervous system expressed only from the maternal allele. Silencing of Ube3a on the paternal allele is thought to occur through the Ube3a-ATS part of LNCAT, since non-coding antisense transcripts are often found at imprinted loci. The deletion and/or mutation of Ube3a on the maternal chromosome causes Angelman Syndrome (AS) and Ube3a-ATS may prove to be an important aspect in finding a therapy for this disease. While in patients with AS the maternal Ube3a allele is inactive, the paternal allele is intact but epigenetically silenced. If unsilenced, the paternal allele could be a source of active Ube3a protein in AS patients. Therefore, understanding the mechanisms of how Ube3a-ATS might be involved in silencing the paternal Ube3a may lead to new therapies for AS. This possibility has been demonstrated by a recent study where the drug topotecan, administered to mice suffering from AS, activated expression of the paternal Ube3a gene by lowering the transcription of Ube3a-ATS.



















