Juvenile hormones (JHs) are a group of acyclic sesquiterpenoids that regulate many aspects of insect physiology. The first discovery of a JH was by Vincent Wigglesworth. JHs regulate development, reproduction, diapause, and polyphenisms. The chemical formula for juvenile hormone is .

The mevalonate pathway, also known as the isoprenoid pathway or HMG-CoA reductase pathway is an essential metabolic pathway present in eukaryotes, archaea, and some bacteria. The pathway produces two five-carbon building blocks called isopentenyl pyrophosphate (IPP) and dimethylallyl pyrophosphate (DMAPP), which are used to make isoprenoids, a diverse class of over 30,000 biomolecules such as cholesterol, vitamin K, coenzyme Q10, and all steroid hormones.

Pyridoxal phosphate (PLP, pyridoxal 5'-phosphate, P5P), the active form of vitamin B6, is a coenzyme in a variety of enzymatic reactions. The International Union of Biochemistry and Molecular Biology has catalogued more than 140 PLP-dependent activities, corresponding to ~4% of all classified activities. The versatility of PLP arises from its ability to covalently bind the substrate, and then to act as an electrophilic catalyst, thereby stabilizing different types of carbanionic reaction intermediates.

Prenylation is the addition of hydrophobic molecules to a protein or a biomolecule. It is usually assumed that prenyl groups (3-methylbut-2-en-1-yl) facilitate attachment to cell membranes, similar to lipid anchors like the GPI anchor, though direct evidence of this has not been observed. Prenyl groups have been shown to be important for protein–protein binding through specialized prenyl-binding domains.
Dolichol refers to any of a group of long-chain mostly unsaturated organic compounds that are made up of varying numbers of isoprene units terminating in an α-saturated isoprenoid group, containing an alcohol functional group.
In molecular biology, biosynthesis is a multi-step, enzyme-catalyzed process where substrates are converted into more complex products in living organisms. In biosynthesis, simple compounds are modified, converted into other compounds, or joined to form macromolecules. This process often consists of metabolic pathways. Some of these biosynthetic pathways are located within a single cellular organelle, while others involve enzymes that are located within multiple cellular organelles. Examples of these biosynthetic pathways include the production of lipid membrane components and nucleotides. Biosynthesis is usually synonymous with anabolism.

Dimethylallyl pyrophosphate is an isoprenoid precursor. It is a product of both the mevalonate pathway and the MEP pathway of isoprenoid precursor biosynthesis. It is an isomer of isopentenyl pyrophosphate (IPP) and exists in virtually all life forms. The enzyme isopentenyl pyrophosphate isomerase catalyzes isomerization between DMAPP and IPP.

Isopentenyl pyrophosphate is an isoprenoid precursor. IPP is an intermediate in the classical, HMG-CoA reductase pathway and in the non-mevalonate MEP pathway of isoprenoid precursor biosynthesis. Isoprenoid precursors such as IPP, and its isomer DMAPP, are used by organisms in the biosynthesis of terpenes and terpenoids.
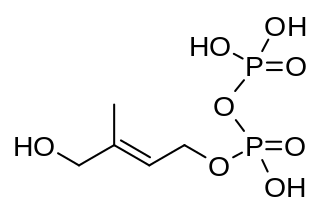
(E)-4-Hydroxy-3-methyl-but-2-enyl pyrophosphate (HMBPP or HMB-PP) is an intermediate of the MEP pathway (non-mevalonate pathway) of isoprenoid biosynthesis. The enzyme HMB-PP synthase (GcpE, IspG) catalyzes the conversion of 2-C-methyl-D-erythritol 2,4-cyclodiphosphate (MEcPP) into HMB-PP. HMB-PP is then converted further to isopentenyl pyrophosphate (IPP) and dimethylallyl pyrophosphate (DMAPP) by HMB-PP reductase (LytB, IspH).
The non-mevalonate pathway—also appearing as the mevalonate-independent pathway and the 2-C-methyl-D-erythritol 4-phosphate/1-deoxy-D-xylulose 5-phosphate (MEP/DOXP) pathway—is an alternative metabolic pathway for the biosynthesis of the isoprenoid precursors isopentenyl pyrophosphate (IPP) and dimethylallyl pyrophosphate (DMAPP). The currently preferred name for this pathway is the MEP pathway, since MEP is the first committed metabolite on the route to IPP.
Squalene synthase (SQS) or farnesyl-diphosphate:farnesyl-diphosphate farnesyl transferase is an enzyme localized to the membrane of the endoplasmic reticulum. SQS participates in the isoprenoid biosynthetic pathway, catalyzing a two-step reaction in which two identical molecules of farnesyl pyrophosphate (FPP) are converted into squalene, with the consumption of NADPH. Catalysis by SQS is the first committed step in sterol synthesis, since the squalene produced is converted exclusively into various sterols, such as cholesterol, via a complex, multi-step pathway. SQS belongs to squalene/phytoene synthase family of proteins.

Isopentenyl pyrophosphate isomerase, also known as Isopentenyl-diphosphate delta isomerase, is an isomerase that catalyzes the conversion of the relatively un-reactive isopentenyl pyrophosphate (IPP) to the more-reactive electrophile dimethylallyl pyrophosphate (DMAPP). This isomerization is a key step in the biosynthesis of isoprenoids through the mevalonate pathway and the MEP pathway.

Mevalonate kinase is an enzyme that in humans is encoded by the MVK gene. Mevalonate kinases are found in a wide variety of organisms from bacteria to mammals. This enzyme catalyzes the following reaction:
ATP citrate synthase (also ATP citrate lyase (ACLY)) is an enzyme that in animals represents an important step in fatty acid biosynthesis. By converting citrate to acetyl-CoA, the enzyme links carbohydrate metabolism, which yields citrate as an intermediate, with fatty acid biosynthesis, which consumes acetyl-CoA. In plants, ATP citrate lyase generates cytosolic acetyl-CoA precursors of thousands of specialized metabolites, including waxes, sterols, and polyketides.

In molecular biology, hydroxymethylglutaryl-CoA synthase or HMG-CoA synthase EC 2.3.3.10 is an enzyme which catalyzes the reaction in which acetyl-CoA condenses with acetoacetyl-CoA to form 3-hydroxy-3-methylglutaryl-CoA (HMG-CoA). This reaction comprises the second step in the mevalonate-dependent isoprenoid biosynthesis pathway. HMG-CoA is an intermediate in both cholesterol synthesis and ketogenesis. This reaction is overactivated in patients with diabetes mellitus type 1 if left untreated, due to prolonged insulin deficiency and the exhaustion of substrates for gluconeogenesis and the TCA cycle, notably oxaloacetate. This results in shunting of excess acetyl-CoA into the ketone synthesis pathway via HMG-CoA, leading to the development of diabetic ketoacidosis.
In enzymology, a 1-deoxy-d-xylulose-5-phosphate synthase (EC 2.2.1.7) is an enzyme in the non-mevalonate pathway that catalyzes the chemical reaction
In enzymology, a geranyltranstransferase is an enzyme that catalyzes the chemical reaction
4-Hydroxy-3-methylbut-2-enyl diphosphate reductase (EC 1.17.1.2, isopentenyl-diphosphate:NADP+ oxidoreductase, LytB, (E)-4-hydroxy-3-methylbut-2-en-1-yl diphosphate reductase, HMBPP reductase, IspH, LytB/IspH) is an enzyme in the non-mevalonate pathway. It acts upon (E)-4-Hydroxy-3-methyl-but-2-enyl pyrophosphate (or "HMB-PP").

Aucubin is an iridoid glycoside. Iridoids are commonly found in plants and function as defensive compounds. Iridoids decrease the growth rates of many generalist herbivores.
Isopentenyl phosphate kinase is an enzyme with systematic name ATP:isopentenyl phosphate phosphotransferase. This enzyme catalyses the following chemical reaction
















