This page is based on this
Wikipedia article Text is available under the
CC BY-SA 4.0 license; additional terms may apply.
Images, videos and audio are available under their respective licenses.

In molecular biology, DNA replication is the biological process of producing two identical replicas of DNA from one original DNA molecule. DNA replication occurs in all living organisms acting as the basis for biological inheritance. The cell possesses the distinctive property of division, which makes replication of DNA essential.

DNA polymerase is an enzyme that synthesizes DNA molecules from deoxyribonucleotides, the building blocks of DNA. These enzymes are essential for DNA replication and usually work in pairs to create two identical DNA strands from a single original DNA molecule. During this process, DNA polymerase "reads" the existing DNA strands to create two new strands that match the existing ones.

DNA polymerase III holoenzyme is the primary enzyme complex involved in prokaryotic DNA replication. It was discovered by Thomas Kornberg and Malcolm Gefter in 1970. The complex has high processivity and, specifically referring to the replication of the E.coli genome, works in conjunction with four other DNA polymerases. Being the primary holoenzyme involved in replication activity, the DNA Pol III holoenzyme also has proofreading capabilities that corrects replication mistakes by means of exonuclease activity working 3'→5'. DNA Pol III is a component of the replisome, which is located at the replication fork.
In molecular biology and biochemistry, processivity is an enzyme's ability to catalyze "consecutive reactions without releasing its substrate".

Okazaki fragments are short sequences of DNA nucleotides which are synthesized discontinuously and later linked together by the enzyme DNA ligase to create the lagging strand during DNA replication. They were discovered in the 1960s by the Japanese molecular biologists Reiji and Tsuneko Okazaki, along with the help of some of their colleagues.

Bolted joints are one of the most common elements in construction and machine design. They consist of fasteners that capture and join other parts, and are secured with the mating of screw threads.
The replication factor C, or RFC, is a five-subunit protein complex that is required for DNA replication.

The replisome is a complex molecular machine that carries out replication of DNA. The replisome first unwinds double stranded DNA into two single strands. For each of the resulting single strands, a new complementary sequence of DNA is synthesized. The net result is formation of two new double stranded DNA sequences that are exact copies of the original double stranded DNA sequence.
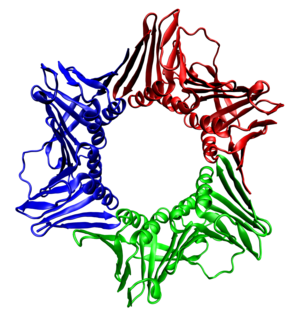
Proliferating cell nuclear antigen (PCNA) is a DNA clamp that acts as a processivity factor for DNA polymerase δ in eukaryotic cells and is essential for replication. PCNA is a homotrimer and achieves its processivity by encircling the DNA, where it acts as a scaffold to recruit proteins involved in DNA replication, DNA repair, chromatin remodeling and epigenetics.

A clamper is an electronic circuit that fixes either the positive or the negative peak excursions of a signal to a defined value by shifting its DC value. The clamper does not restrict the peak-to-peak excursion of the signal, it moves the whole signal up or down so as to place the peaks at the reference level. A diode clamp consists of a diode, which conducts electric current in only one direction and prevents the signal exceeding the reference value; and a capacitor, which provides a DC offset from the stored charge. The capacitor forms a time constant with the resistor load, which determines the range of frequencies over which the clamper will be effective.

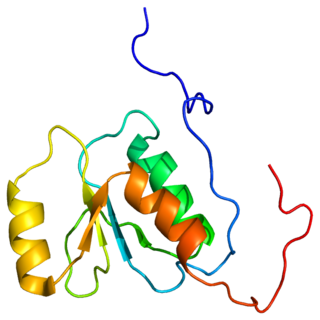
A DNA clamp, also known as a sliding clamp or β-clamp, is a protein fold that serves as a processivity-promoting factor in DNA replication. As a critical component of the DNA polymerase III holoenzyme, the clamp protein binds DNA polymerase and prevents this enzyme from dissociating from the template DNA strand. The clamp-polymerase protein–protein interactions are stronger and more specific than the direct interactions between the polymerase and the template DNA strand; because one of the rate-limiting steps in the DNA synthesis reaction is the association of the polymerase with the DNA template, the presence of the sliding clamp dramatically increases the number of nucleotides that the polymerase can add to the growing strand per association event. The presence of the DNA clamp can increase the rate of DNA synthesis up to 1,000-fold compared with a nonprocessive polymerase.

Eukaryotic DNA replication is a conserved mechanism that restricts DNA replication to once per cell cycle. Eukaryotic DNA replication of chromosomal DNA is central for the duplication of a cell and is necessary for the maintenance of the eukaryotic genome.

Replication factor C subunit 5 is a protein that in humans is encoded by the RFC5 gene.

Chromosome transmission fidelity protein 8 homolog is a protein that in humans is encoded by the CHTF8 gene.
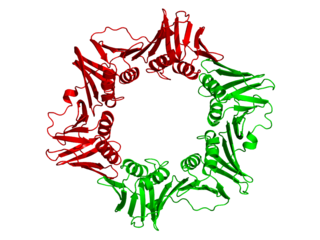
dnaN is the gene that codes for the DNA clamp of DNA polymerase III in prokaryotes. The β clamp physically locks Pol III onto a DNA strand during replication to help increase its processivity. The eukaryotic equivalent to the β clamp is PCNA.

A circular prokaryote chromosome is a chromosome in bacteria and archaea, in the form of a molecule of circular DNA. Unlike the linear DNA of most eukaryotes, typical prokaryote chromosomes are circular.
Sister chromatid cohesion refers to the process by which sister chromatids are paired and held together during certain phases of the cell cycle. Establishment of sister chromatid cohesion is the process by which chromatin-associated cohesin protein becomes competent to physically bind together the sister chromatids. In general, cohesion is established during S phase as DNA is replicated, and is lost when chromosomes segregate during mitosis and meiosis. Some studies have suggested that cohesion aids in aligning the kinetochores during mitosis by forcing the kinetochores to face opposite cell poles.
DNA polymerase delta is an enzyme complex found in eukaryotes that is involved in DNA replication and repair. The DNA polymerase delta complex consists of 4 subunits: POLD1, POLD2, POLD3, and POLD4. DNA Pol δ is an enzyme used for both leading and lagging strand synthesis. It exhibits increased processivity when interacting with the proliferating cell nuclear antigen (PCNA). As well, the multisubunit protein replication factor C, through its role as the clamp loader for PCNA is important for DNA Pol δ function.
DNA polymerase epsilon is a member of the DNA polymerase family of enzymes found in eukaryotes. It is composed of the following four subunits: POLE, POLE2, POLE3, and POLE4. Recent evidence suggests that it plays a major role in leading strand DNA synthesis and nucleotide and base excision repair.