Antiviral drugs are a class of medication used specifically for treating viral infections rather than bacterial ones. Most antivirals are used for specific viral infections, while a broad-spectrum antiviral is effective against a wide range of viruses. Unlike most antibiotics, antiviral drugs do not destroy their target pathogen; instead they inhibit their development.

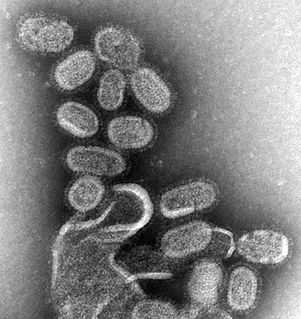
Influenza hemagglutinin (HA) or haemagglutinin[p] is a homotrimeric glycoprotein found on the surface of influenza viruses and integral to its infectivity.
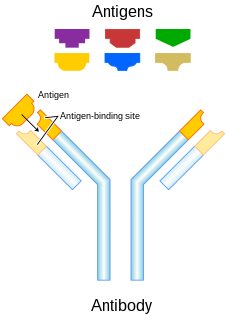
Antigenic drift is a mechanism for variation in viruses that involves the accumulation of mutations within the genes that code for antibody-binding sites. This results in a new strain of virus particles which cannot be inhibited as effectively by the antibodies that were originally targeted against previous strains, making it easier for the virus to spread throughout a partially immune population. Antigenic drift occurs in both influenza A and influenza B viruses.
Rabies lyssavirus, formerly Rabies virus, is a neurotropic virus that causes rabies in humans and animals. Rabies transmission can occur through the saliva of animals and less commonly through contact with human saliva. Rabies lyssavirus, like many rhabdoviruses, has an extremely wide host range. In the wild it has been found infecting many mammalian species, while in the laboratory it has been found that birds can be infected, as well as cell cultures from mammals, birds, reptiles and insects.
Hemagglutinin esterase (HEs) is a glycoprotein that certain enveloped viruses possess and use as invading mechanism. HEs helps in the attachment and destruction of certain sialic acid receptors that are found on the host cell surface. Viruses that possess HEs include influenza C, Toro-viruses, and coronaviruses. HEs is a dimer transmembrane protein consisting of two monomers, each monomer is made of three domains. The three domains are: membrane fusion, esterase, and receptor binding domains.

In biology and immunology, an Alphavirus belongs to group IV of the Baltimore classification of the Togaviridae family of viruses, according to the system of classification based on viral genome composition introduced by David Baltimore in 1971. Alphaviruses, like all other group IV viruses, have a positive sense, single-stranded RNA genome. There are thirty alphaviruses able to infect various vertebrates such as humans, rodents, fish, birds, and larger mammals such as horses as well as invertebrates. Transmission between species and individuals occurs mainly via mosquitoes, making the alphaviruses a member of the collection of arboviruses – or arthropod-borne viruses. Alphavirus particles are enveloped, have a 70 nm diameter, tend to be spherical, and have a 40 nm isometric nucleocapsid.

Thogotovirus is a genus of enveloped RNA viruses, one of seven genera in the virus family Orthomyxoviridae. Their single-stranded, negative-sense RNA genome has six or seven segments. Thogotoviruses are distinguished from most other orthomyxoviruses by being arboviruses – viruses that are transmitted by arthropods, in this case usually ticks. Thogotoviruses can replicate in both tick cells and vertebrate cells; one subtype has also been isolated from mosquitoes. A consequence of being transmitted by blood-sucking vectors is that the virus must spread systemically in the vertebrate host – unlike influenza viruses, which are transmitted by respiratory droplets and are usually confined to the respiratory system.

H5N1 genetic structure is the molecular structure of the H5N1 virus's RNA.
Antigenic variation refers to the mechanism by which an infectious agent such as a protozoan, bacterium or virus alters its surface proteins in order to avoid a host immune response. It is related to phase variation. Immune evasion is particularly important for organisms that target long-lived hosts, repeatedly infect a single host and are easily transmittable. Antigenic variation not only enables immune evasion by the pathogen, but also allows the microbes to cause re-infection, as their antigens are no longer recognized by the host's immune system. When an organism is exposed to a particular antigen an immune response is stimulated and antibodies are generated to target that specific antigen. The immune system will then "remember" that particular antigen, and defenses aimed at that antigen become part of the immune system’s acquired immune response. If the same pathogen tries to re-infect the same host the antibodies will act rapidly to target the pathogen for destruction. However, if the pathogen can alter its surface antigens, it can evade the host's acquired immune system. This will allow the pathogen to re-infect the host while the immune system generates new antibodies to target the newly identified antigen. Antigenic variation can occur by altering a variety of surface molecules including proteins and carbohydrates. There are many molecular mechanisms behind antigenic variation, including gene conversion, site-specific DNA inversions, hypermutation, as well as recombination of sequence cassettes. In all cases, antigenic variation and phase variation result in a heterogenic phenotype of a clonal population. Individual cells either express the phase-variable protein(s) or express one of multiple antigenic forms of the protein. This form of regulation has been identified mainly, but not exclusively, for a wide variety of surface structures in pathogens and is implicated as a virulence strategy.
Sendai virus (SeV), previously also known as murine parainfluenza virus type 1 or hemagglutinating virus of Japan (HVJ), is a negative sense, single-stranded RNA virus of the family Paramyxoviridae, a group of viruses featuring, notably, the genera Morbillivirus and Rubulavirus. SeV is a member of genus Respirovirus, members of which primarily infect mammals.
Env is a viral gene that encodes the protein forming the viral envelope. The expression of the env gene enables retroviruses to target and attach to specific cell types, and to infiltrate the target cell membrane.

A virus is a biological agent that reproduces inside the cells of living hosts. When infected by a virus, a host cell is forced to produce thousands of identical copies of the original virus at an extraordinary rate. Unlike most living things, viruses do not have cells that divide; new viruses are assembled in the infected host cell. But unlike still simpler infectious agents, viruses contain genes, which gives them the ability to mutate and evolve. Over 5,000 species of viruses have been discovered.
Avian sarcoma leukosis virus (ASLV) is an endogenous retrovirus that infects and can lead to cancer in chickens; experimentally it can infect other species of birds and mammals. ASLV replicates in chicken embryo fibroblasts, the cells that contribute to the formation of connective tissues. Different forms of the disease exist, including lymphoblastic, erythroblastic, and osteopetrotic.

Viral neuraminidase is a type of neuraminidase found on the surface of influenza viruses that enables the virus to be released from the host cell. Neuraminidases are enzymes that cleave sialic acid groups from glycoproteins and are required for influenza virus replication.
Virus quantification involves counting the number of viruses in a specific volume to determine the virus concentration. It is utilized in both research and development (R&D) in commercial and academic laboratories as well as production situations where the quantity of virus at various steps is an important variable. For example, the production of viral vaccines, recombinant proteins using viral vectors and viral antigens all require virus quantification to continually adapt and monitor the process in order to optimize production yields and respond to ever changing demands and applications. Examples of specific instances where known viruses need to be quantified include clone screening, multiplicity of infection (MOI) optimization and adaptation of methods to cell culture. This page discusses various techniques currently used to quantify viruses in liquid samples. These methods are separated into two categories, traditional vs. modern methods. Traditional methods are industry-standard methods that have been used for decades but are generally slow and labor-intensive. Modern methods are relatively new commercially available products and kits that greatly reduce quantification time. This is not meant to be an exhaustive review of all potential methods, but rather a representative cross-section of traditional methods and new, commercially available methods. While other published methods may exist for virus quantification, non-commercial methods are not discussed here.

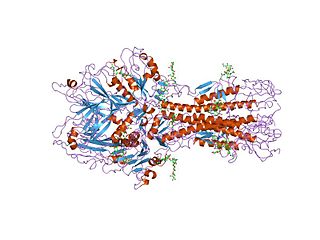
In molecular biology, haemagglutinin-esterase fusion glycoprotein (HEF) is a multi-functional protein embedded in the viral envelope of several viruses, including influenza C virus, influenza D virus, coronaviruses, and toroviruses. HEF is required for infectivity, and functions to recognise the host cell surface receptor, to fuse the viral and host cell membranes, and to destroy the receptor upon host cell infection. The haemagglutinin region of HEF is responsible for receptor recognition and membrane fusion, and bears a strong resemblance to the sialic acid-binding haemagglutinin found in influenza A and B viruses, except that it binds 9-O-acetylsialic acid. The esterase region of HEF is responsible for the destruction of the receptor, an action that is carried out by neuraminidase in influenza A and B viruses. The esterase domain is similar in structure to Streptomyces scabies esterase, and to acetylhydrolase, thioesterase I and rhamnogalacturonan acetylesterase.
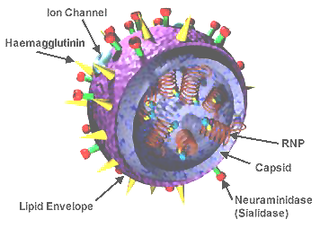
Influenza D virus is a species in the virus genus Influenzavirus D in the family Orthomyxoviridae, that causes influenza.