
The hepatitis C virus (HCV) is a small, enveloped, positive-sense single-stranded RNA virus of the family Flaviviridae. The hepatitis C virus is the cause of hepatitis C and some cancers such as liver cancer and lymphomas in humans.
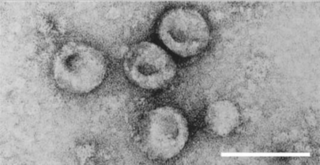
Pestivirus is a genus of viruses, in the family Flaviviridae. Viruses in the genus Pestivirus infect mammals, including members of the family Bovidae and the family Suidae. There are 11 species in this genus. Diseases associated with this genus include: hemorrhagic syndromes, abortion, and fatal mucosal disease.
The NS1 influenza protein (NS1) is a viral nonstructural protein encoded by the NS gene segments of type A, B and C influenza viruses. Also encoded by this segment is the nuclear export protein (NEP), formally referred to as NS2 protein, which mediates the export of influenza virus ribonucleoprotein (RNP) complexes from the nucleus, where they are assembled.

Murine coronavirus (M-CoV) is a virus in the genus Betacoronavirus that infects mice. Belonging to the subgenus Embecovirus, murine coronavirus strains are enterotropic or polytropic. Enterotropic strains include mouse hepatitis virus (MHV) strains D, Y, RI, and DVIM, whereas polytropic strains, such as JHM and A59, primarily cause hepatitis, enteritis, and encephalitis. Murine coronavirus is an important pathogen in the laboratory mouse and the laboratory rat. It is the most studied coronavirus in animals other than humans, and has been used as an animal disease model for many virological and clinical studies.

The Coronavirus packaging signal is a conserved cis-regulatory element found in Betacoronavirus. It has an important role in regulating the packaging of the viral genome into the capsid. As part of the viral life cycle, within the infected cell, the viral genome becomes associated with viral proteins and assembles into new infective progeny viruses. This process is called packaging and is vital for viral replication.
The Hepatitis C stem-loop IV is part of a putative RNA element found in the NS5B coding region. This element along with stem-loop VII, is important for colony formation, though its exact function and mechanism are unknown.

The hepatitis C virus 3′X element is an RNA element which contains three stem-loop structures that are essential for replication.

The Hepatitis C virus (HCV) cis-acting replication element (CRE) is an RNA element which is found in the coding region of the RNA-dependent RNA polymerase NS5B. Mutations in this family have been found to cause a blockage in RNA replication and it is thought that both the primary sequence and the structure of this element are crucial for HCV RNA replication.
Hepatitis C virus stem-loop VII is a regulatory element found in the coding region of the RNA-dependent RNA polymerase gene, NS5B. Similarly to stem-loop IV, the stem-loop structure is important for colony formation, though its exact function and mechanism are unknown.
NSP1 (NS53), the product of rotavirus gene 5, is a nonstructural RNA-binding protein that contains a cysteine-rich region and is a component of early replication intermediates. RNA-folding predictions suggest that this region of the NSP1 mRNA can interact with itself, producing a stem-loop structure similar to that found near the 5'-terminus of the NSP1 mRNA.

NSP2 (NS35), is a rotavirus nonstructural RNA-binding protein that accumulates in cytoplasmic inclusions (viroplasms) and is required for genome replication. NSP2 is closely associated in vivo with the viral replicase. The non-structural protein NSP5 plays a role in the structure of viroplasms mediated by its interaction with NSP2.

VAMP-Associated Protein A is a protein that in humans is encoded by the VAPA gene. Together with VAPB and VAPC it forms the VAP protein family. They are integral endoplasmic reticulum membrane proteins of the type II and are ubiquitous among eukaryotes.

Nonstructural protein 3 (NS3), also known as p-70, is a viral nonstructural protein that is 70 kDa cleavage product of the hepatitis C virus polyprotein. It acts as a serine protease. C-terminal two-thirds of the protein also acts as helicase and nucleoside triphosphatase. First (N-terminal) 180 aminoacids of NS3 has additional role as cofactor domains for NS2 protein.

Nonstructural protein 2 (NS2) is a viral protein found in the hepatitis C virus. It is also produced by influenza viruses, and is alternatively known as the nuclear export protein (NEP).

HBx is a hepatitis B viral protein. It is 154 amino acids long and interferes with transcription, signal transduction, cell cycle progress, protein degradation, apoptosis and chromosomal stability in the host. It forms a heterodimeric complex with its cellular target protein, and this interaction dysregulates centrosome dynamics and mitotic spindle formation. It interacts with DDB1 redirecting the ubiquitin ligase activity of the CUL4-DDB1 E3 complexes, which are intimately involved in the intracellular regulation of DNA replication and repair, transcription and signal transduction.
RIG-I-like receptors are a type of intracellular pattern recognition receptor involved in the recognition of viruses by the innate immune system. RIG-I is the best characterized receptor within the RIG-I like receptor (RLR) family. Together with MDA5 and LGP2, this family of cytoplasmic pattern recognition receptors (PRRs) are sentinels for intracellular viral RNA that is a product of viral infection. The RLR receptors provide frontline defence against viral infections in most tissues.

Daclatasvir, sold under the brand name Daklinza, is an antiviral medication used in combination with other medications to treat hepatitis C (HCV). The other medications used in combination include sofosbuvir, ribavirin, and interferon, vary depending on the virus type and whether the person has cirrhosis. It is taken by mouth.

Minute virus of mice (MVM) is the exemplar virus of the species Rodent protoparvovirus 1, in the genus Protoparvovirus of the Parvoviridae family of viruses. MVM exists in multiple variant forms including MVMp, which is the prototype strain that infects cells of fibroblast origin, while MVMi, the immunosuppressive strain, infects T lymphocytes. MVM is a common infection in laboratory mice due to its highly contagious nature. The virus can be shed from infected mice via feces and urine, but also via fomites and nasal secretions. Typically there are no clinical signs of infection in adult mice, however, experimental infection can cause multiple organ damage during fetal development or shortly after birth.

Nonstructural protein 5B (NS5B) is a viral protein found in the hepatitis C virus (HCV). It is an RNA-dependent RNA polymerase, having the key function of replicating HCV's viral RNA by using the viral positive RNA strand as a template to catalyze the polymerization of ribonucleoside triphosphates (rNTP) during RNA replication. Several crystal structures of NS5B polymerase in several crystalline forms have been determined based on the same consensus sequence BK. The structure can be represented by a right hand shape with fingers, palm, and thumb. The encircled active site, unique to NS5B, is contained within the palm structure of the protein. Recent studies on NS5B protein genotype 1b strain J4's (HC-J4) structure indicate a presence of an active site where possible control of nucleotide binding occurs and initiation of de-novo RNA synthesis. De-novo adds necessary primers for initiation of RNA replication.

Nonstructural protein 5A (NS5A) inhibitors are direct acting antiviral agents (DAAs) that target viral proteins, and their development was a culmination of increased understanding of the viral life cycle combined with advances in drug discovery technology. However, their mechanism of action is complex and not fully understood. NS5A inhibitors were the focus of much attention when they emerged as a part of the first curative treatment for hepatitis C virus (HCV) infections in 2014. Favorable characteristics have been introduced through varied structural changes, and structural similarities between NS5A inhibitors that are clinically approved are readily apparent. Despite the recent introduction of numerous new antiviral drugs, resistance is still a concern and these inhibitors are therefore always used in combination with other drugs.














