Hepatitis D is a type of viral hepatitis caused by the hepatitis delta virus (HDV). HDV is one of five known hepatitis viruses: A, B, C, D, and E. HDV is considered to be a satellite because it can propagate only in the presence of the hepatitis B virus (HBV). Transmission of HDV can occur either via simultaneous infection with HBV (coinfection) or superimposed on chronic hepatitis B or hepatitis B carrier state (superinfection).
Reverse-transcriptase inhibitors (RTIs) are a class of antiretroviral drugs used to treat HIV infection or AIDS, and in some cases hepatitis B. RTIs inhibit activity of reverse transcriptase, a viral DNA polymerase that is required for replication of HIV and other retroviruses.

The hepatitis C virus (HCV) is a small, enveloped, positive-sense single-stranded RNA virus of the family Flaviviridae. The hepatitis C virus is the cause of hepatitis C and some cancers such as liver cancer and lymphomas in humans.

RNA-dependent RNA polymerase (RdRp) or RNA replicase is an enzyme that catalyzes the replication of RNA from an RNA template. Specifically, it catalyzes synthesis of the RNA strand complementary to a given RNA template. This is in contrast to typical DNA-dependent RNA polymerases, which all organisms use to catalyze the transcription of RNA from a DNA template.

This family represents the internal ribosome entry site (IRES) of the hepatitis A virus. HAV IRES is a 450 nucleotide long sequence located in the 735 nt long 5’ UTR of Hepatitis A viral RNA genome. IRES elements allow cap and end-independent translation of mRNA in the host cell. The IRES achieves this by mediating the internal initiation of translation by recruiting a ribosomal 40S pre-initiation complex directly to the initiation codon and eliminates the requirement for eukaryotic initiation factor, eIF4F.

The Hepatitis C virus (HCV) cis-acting replication element (CRE) is an RNA element which is found in the coding region of the RNA-dependent RNA polymerase NS5B. Mutations in this family have been found to cause a blockage in RNA replication and it is thought that both the primary sequence and the structure of this element are crucial for HCV RNA replication.
NS2-3 protease is an enzyme responsible for proteolytic cleavage between NS2 and NS3, which are non-structural proteins that form part of the HCV virus particle. NS3 protease of hepatitis C virus, on the other hand, is responsible for the cleavage of non-structural protein downstream. Both of these proteases are directly involved in HCV genome replication, that is, during the viral life-cycle that leads to virus multiplication in the host that has been infected by the virus.

Nonstructural protein 3 (NS3), also known as p-70, is a viral nonstructural protein that is 70 kDa cleavage product of the hepatitis C virus polyprotein. It acts as a serine protease. C-terminal two-thirds of the protein also acts as helicase and nucleoside triphosphatase. First (N-terminal) 180 aminoacids of NS3 has additional role as cofactor domains for NS2 protein.

Nonstructural protein 5A (NS5A) is a zinc-binding and proline-rich hydrophilic phosphoprotein that plays a key role in Hepatitis C virus RNA replication. It appears to be a dimeric form without trans-membrane helices.

Nonstructural protein 2 (NS2) is a viral protein found in the hepatitis C virus. It is also produced by influenza viruses, and is alternatively known as the nuclear export protein (NEP).

Hepatitis B virus DNA polymerase is a hepatitis B viral protein. It is a DNA polymerase that can use either DNA or RNA templates and a ribonuclease H that cuts RNA in the duplex. Both functions are supplied by the reverse transcriptase (RT) domain.

Hepatitis B virus (HBV) is a partially double-stranded DNA virus, a species of the genus Orthohepadnavirus and a member of the Hepadnaviridae family of viruses. This virus causes the disease hepatitis B.

Daclatasvir, sold under the brand name Daklinza, is an antiviral medication used in combination with other medications to treat hepatitis C (HCV). The other medications used in combination include sofosbuvir, ribavirin, and interferon, vary depending on the virus type and whether the person has cirrhosis. It is taken by mouth.

Ledipasvir is a drug for the treatment of hepatitis C that was developed by Gilead Sciences. After completing Phase III clinical trials, on February 10, 2014, Gilead filed for U.S. approval of a ledipasvir/sofosbuvir fixed-dose combination tablet for genotype 1 hepatitis C. The ledipasvir/sofosbuvir combination is a direct-acting antiviral agent that interferes with HCV replication and can be used to treat patients with genotypes 1a or 1b without PEG-interferon or ribavirin.

Ledipasvir/sofosbuvir, sold under the trade name Harvoni among others, is a medication used to treat hepatitis C. It is a fixed-dose combination of ledipasvir and sofosbuvir. Cure rates are 94% to 99% in people infected with hepatitis C virus (HCV) genotype 1. Some evidence also supports use in HCV genotype 3 and 4. It is taken daily by mouth for 8–24 weeks.
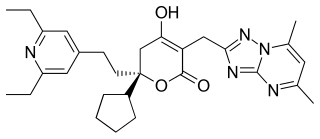
Filibuvir was a non-nucleoside orally available NS5B inhibitor developed by Pfizer for the treatment of hepatitis C. It binds to the non-catalytic Thumb II allosteric pocket of NS5B viral polymerase and causes a decrease in viral RNA synthesis. It is a potent and selective inhibitor, with a mean IC50 of 0.019 μM against genotype 1 polymerases. Several filibuvir-resistant mutations have been identified, M423 being the most common that occurred after filibuvir monotherapy. It was intended to be taken twice-daily.

Nonstructural protein 5A (NS5A) inhibitors are direct acting antiviral agents (DAAs) that target viral proteins, and their development was a culmination of increased understanding of the viral life cycle combined with advances in drug discovery technology. However, their mechanism of action is complex and not fully understood. NS5A inhibitors were the focus of much attention when they emerged as a part of the first curative treatment for hepatitis C virus (HCV) infections in 2014. Favorable characteristics have been introduced through varied structural changes, and structural similarities between NS5A inhibitors that are clinically approved are readily apparent. Despite the recent introduction of numerous new antiviral drugs, resistance is still a concern and these inhibitors are therefore always used in combination with other drugs.
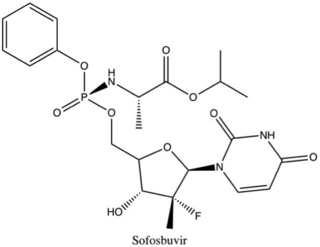
Non-structural protein 5B (NS5B) inhibitors are a class of direct-acting antivirals widely used in the treatment of chronic hepatitis C. Depending on site of action and chemical composition, NS5B inhibitors may be categorized into three classes—nucleoside active site inhibitors (NIs), non-nucleoside allosteric inhibitors, and pyrophosphate analogues. Subsequently, all three classes are then subclassified. All inhibit RNA synthesis by NS5B but at different stages/sites resulting in inability of viral RNA replication. Expression of direct-acting NS5B inhibitors does not take place in cells that are not infected by hepatitis C virus, which seems to be beneficial for this class of drugs.
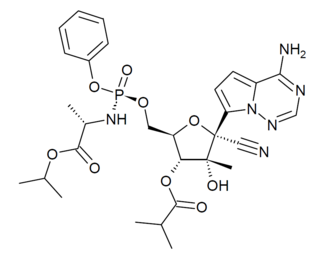
GS-6620 is an antiviral drug which is a nucleotide analogue. It was developed for the treatment of Hepatitis C but while it showed potent antiviral effects in early testing, it could not be successfully formulated into an oral dosage form due to low and variable absorption in the intestines which made blood levels unpredictable. It has however continued to be researched as a potential treatment for other viral diseases such as Ebola virus disease.
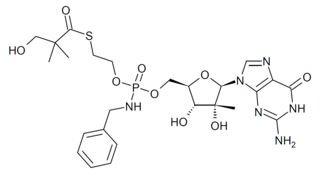
IDX-184 is an antiviral drug which was developed as a treatment for hepatitis C, acting as a NS5B RNA polymerase inhibitor. While it showed reasonable effectiveness in early clinical trials it did not progress past Phase IIb. However research using this drug has continued as it shows potentially useful activity against other emerging viral diseases such as Zika virus, and coronaviruses including MERS, and SARS-CoV-2.













