
Nucleotides are organic molecules composed of a nitrogenous base, a pentose sugar and a phosphate. They serve as monomeric units of the nucleic acid polymers – deoxyribonucleic acid (DNA) and ribonucleic acid (RNA), both of which are essential biomolecules within all life-forms on Earth. Nucleotides are obtained in the diet and are also synthesized from common nutrients by the liver.
A nucleoside triphosphate is a nucleoside containing a nitrogenous base bound to a 5-carbon sugar, with three phosphate groups bound to the sugar. They are the molecular precursors of both DNA and RNA, which are chains of nucleotides made through the processes of DNA replication and transcription. Nucleoside triphosphates also serve as a source of energy for cellular reactions and are involved in signalling pathways.

Nucleoside-diphosphate kinases are enzymes that catalyze the exchange of terminal phosphate between different nucleoside diphosphates (NDP) and triphosphates (NTP) in a reversible manner to produce nucleotide triphosphates. Many NDP serve as acceptor while NTP are donors of phosphate group. The general reaction via ping-pong mechanism is as follows: XDP + YTP ←→ XTP + YDP. NDPK activities maintain an equilibrium between the concentrations of different nucleoside triphosphates such as, for example, when guanosine triphosphate (GTP) produced in the citric acid (Krebs) cycle is converted to adenosine triphosphate (ATP). Other activities include cell proliferation, differentiation and development, signal transduction, G protein-coupled receptor, endocytosis, and gene expression.

Apyrase is a calcium-activated plasma membrane-bound enzyme that catalyses the hydrolysis of ATP to yield AMP and inorganic phosphate. Two isoenzymes are found in commercial preparations from S. tuberosum. One with a higher ratio of substrate selectivity for ATP:ADP and another with no selectivity.
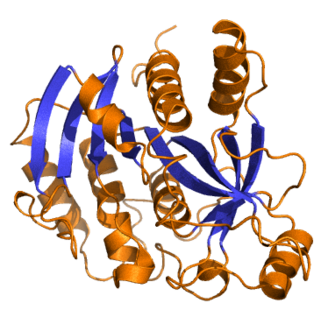
Purine nucleoside phosphorylase, PNP, PNPase or inosine phosphorylase is an enzyme that in humans is encoded by the NP gene. It catalyzes the chemical reaction

Nucleic acid metabolism is a collective term that refers to the variety of chemical reactions by which nucleic acids are either synthesized or degraded. Nucleic acids are polymers made up of a variety of monomers called nucleotides. Nucleotide synthesis is an anabolic mechanism generally involving the chemical reaction of phosphate, pentose sugar, and a nitrogenous base. Degradation of nucleic acids is a catabolic reaction and the resulting parts of the nucleotides or nucleobases can be salvaged to recreate new nucleotides. Both synthesis and degradation reactions require multiple enzymes to facilitate the event. Defects or deficiencies in these enzymes can lead to a variety of diseases.
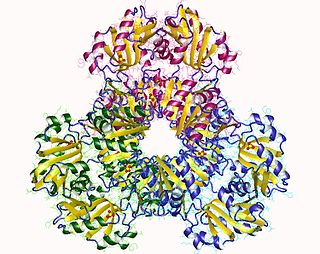
Ribose-phosphate diphosphokinase is an enzyme that converts ribose 5-phosphate into phosphoribosyl pyrophosphate (PRPP). It is classified under EC 2.7.6.1.
In enzymology, a 6-carboxyhexanoate—CoA ligase is an enzyme that catalyzes the chemical reaction
In enzymology, a dethiobiotin synthase (EC 6.3.3.3) is an enzyme that catalyzes the chemical reaction
In enzymology, a glutamate—ethylamine ligase (EC 6.3.1.6) is an enzyme that catalyzes the chemical reaction
In enzymology, an acetoin-ribose-5-phosphate transaldolase is an enzyme that catalyzes the chemical reaction
In enzymology, an adenosylmethionine-8-amino-7-oxononanoate transaminase is an enzyme that catalyzes the chemical reaction

In enzymology, a 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine diphosphokinase is an enzyme that catalyzes the chemical reaction
In enzymology, a GTP diphosphokinase is an enzyme that catalyzes the chemical reaction
In enzymology, a nucleoside-triphosphate-adenylate kinase is an enzyme that catalyzes the chemical reaction
In enzymology, a thiamine diphosphokinase is an enzyme that catalyzes the chemical reaction
Nucleoside oxidase (EC 1.1.3.28) is an enzyme with systematic name nucleoside:oxygen 5'-oxidoreductase. This enzyme catalyses the following chemical reaction
Rhodotorulapepsin is an enzyme. This enzyme catalyses the following chemical reaction
Scytalidopepsin A (EC 3.4.23.31, Scytalidium aspartic proteinase A, Scytalidium lignicolum aspartic proteinase, Scytalidium lignicolum aspartic proteinase A-2, Scytalidium lignicolum aspartic proteinase A-I, Scytalidium lignicolum aspartic proteinase C, Scytalidium lignicolum carboxyl proteinase, Scytalidium lignicolum acid proteinase) is an enzyme. This enzyme catalyses the following chemical reaction
Jun-Ichiro Mukai is a Japanese biochemist, emeritus professor at faculty of agriculture, Kyushu University.







