
Glucose-6-phosphate dehydrogenase (G6PD or G6PDH) (EC 1.1.1.49) is a cytosolic enzyme that catalyzes the chemical reaction
Purine metabolism refers to the metabolic pathways to synthesize and break down purines that are present in many organisms.
In enzymology, a (R)-2-hydroxyacid dehydrogenase (EC 1.1.1.272) is an enzyme that catalyzes the chemical reaction
In enzymology, a coenzyme F420 hydrogenase (EC 1.12.98.1) is an enzyme that catalyzes the chemical reaction
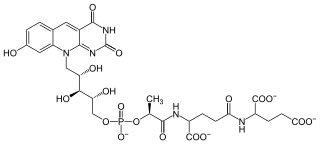
Coenzyme F420 or 8-hydroxy-5-deazaflavin is a coenzyme (sometimes called a cofactor) involved in redox reactions in methanogens, in many Actinomycetota, and sporadically in other bacterial lineages. It is a flavin derivative with an absorption maximum at 420 nm—hence its name. The coenzyme is a substrate for coenzyme F420 hydrogenase, 5,10-methylenetetrahydromethanopterin reductase and methylenetetrahydromethanopterin dehydrogenase.

In molecular biology, hydroxymethylglutaryl-CoA synthase or HMG-CoA synthase EC 2.3.3.10 is an enzyme which catalyzes the reaction in which acetyl-CoA condenses with acetoacetyl-CoA to form 3-hydroxy-3-methylglutaryl-CoA (HMG-CoA). This reaction comprises the second step in the mevalonate-dependent isoprenoid biosynthesis pathway. HMG-CoA is an intermediate in both cholesterol synthesis and ketogenesis. This reaction is overactivated in patients with diabetes mellitus type 1 if left untreated, due to prolonged insulin deficiency and the exhaustion of substrates for gluconeogenesis and the TCA cycle, notably oxaloacetate. This results in shunting of excess acetyl-CoA into the ketone synthesis pathway via HMG-CoA, leading to the development of diabetic ketoacidosis.

In enzymology, a 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine diphosphokinase is an enzyme that catalyzes the chemical reaction
In enzymology, a 4-(cytidine 5'-diphospho)-2-C-methyl-D-erythritol kinase is an enzyme that catalyzes the chemical reaction
2,5-diamino-6-(ribosylamino)-4(3H)-pyrimidinone 5'-phosphate reductase (EC 1.1.1.302, 2,5-diamino-6-ribosylamino-4(3H)-pyrimidinone 5'-phosphate reductase, MjaRED, MJ0671 (gene)) is an enzyme with systematic name 2,5-diamino-6-(5-phospho-D-ribosylamino)pyrimidin-4(3H)-one:NAD(P)+ oxidoreductase. This enzyme catalyses the following chemical reaction

L-2-hydroxycarboxylate dehydrogenase (NAD+) (EC 1.1.1.337, (R)-sulfolactate:NAD+ oxidoreductase, L-sulfolactate dehydrogenase, (R)-sulfolactate dehydrogenase, L-2-hydroxyacid dehydrogenase (NAD+), ComC) is an enzyme with systematic name (2S)-2-hydroxycarboxylate:NAD+ oxidoreductase. This enzyme catalyses the following chemical reaction
7,8-didemethyl-8-hydroxy-5-deazariboflavin synthase (EC 4.3.1.32, FO synthase) and 5-amino-6-(D-ribitylamino)uracil—L-tyrosine 4-hydroxyphenyl transferase (EC 2.5.1.147) are two enzymes always complexed together to achieve synthesis of FO, a precursor to Coenzyme F420. Their systematic names are 5-amino-5-(4-hydroxybenzyl)-6-(D-ribitylimino)-5,6-dihydrouracil ammonia-lyase (7,8-didemethyl-8-hydroxy-5-deazariboflavin-forming) and 5-amino-6-(D-ribitylamino)uracil:L-tyrosine, 4-hydroxyphenyl transferase respectively. The enzymes catalyse the following chemical reactions:
Phosphoserine transaminase is an enzyme with systematic name O-phospho-L-serine:2-oxoglutarate aminotransferase. This enzyme catalyses the following chemical reaction
2-Phospho-L-lactate guanylyltransferase is an enzyme with systematic name GTP:2-phospho-L-lactate guanylyltransferase. This enzyme catalyses the following chemical reaction
2-amino-5-formylamino-6-ribosylaminopyrimidin-4(3H)-one 5'-monophosphate deformylase (EC 3.5.1.102, ArfB) is an enzyme with systematic name 2-amino-5-formylamino-6-(5-phospho-D-ribosylamino)pyrimidin-4(3H)-one amidohydrolase. This enzyme catalyses the following chemical reaction
Coenzyme F420-0:L-glutamate ligase (EC 6.3.2.31, CofE-AF, MJ0768, CofE) is an enzyme with systematic name L-glutamate:coenzyme F420-0 ligase (GDP-forming). This enzyme catalyses the following chemical reaction
Coenzyme gamma-F420-2:α-L-glutamate ligase (EC 6.3.2.32, MJ1001, CofF protein, gamma-F420-2:alpha-L-glutamate ligase) is an enzyme with systematic name L-glutamate:coenzyme gamma-F420-2 (ADP-forming). This enzyme catalyses the following chemical reaction
Tetrahydrosarcinapterin synthase is an enzyme with systematic name tetrahydromethanopterin:alpha-L-glutamate ligase (ADP-forming). This enzyme catalyses the following chemical reaction
Coenzyme F420-1:γ-L-glutamate ligase (EC 6.3.2.34, F420:gamma-glutamyl ligase, CofE-AF, MJ0768, CofE) is an enzyme with systematic name L-glutamate:coenzyme F420-1 ligase (GDP-forming). This enzyme catalyses the following chemical reaction

2,5-diamino-6-hydroxy-4-(5-phosphoribosylamino)pyrimidine is a metabolite in the purine metabolism, formed by the hydrolysis of GTP by GTP cyclohydrolase II. Alternatively two separate enzymes can carry out this reaction, initially GTP cyclohydrolase IIa hydrolyses the 8,9 bond to form 2-Amino-5-formylamino-6-(5-phospho-D-ribosylamino)pyrimidin-4(3H)-one, followed by de-formylation by 2-amino-5-formylamino-6-ribosylaminopyrimidin-4(3H)-one 5'-monophosphate deformylase. 2,5-diamino-6-hydroxy-4-(5-phosphoribosylamino)pyrimidine is deaminated by Diaminohydroxyphosphoribosylaminopyrimidine deaminase to form 5-amino-6-(5-phosphoribosylamino)uracil.
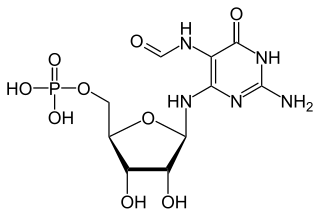
2-Amino-5-formylamino-6-(5-phospho-D-ribosylamino)pyrimidin-4(3H)-one is a metabolite in the riboflavin biosynthesis pathway. It is formed from GTP by the enzyme GTP cyclohydrolase IIa which catalyzes the hydrolysis of the 8,9 bond in the guanine group and loss of the beta and gamma phosphate groups. The molecule is deformylated by 2-amino-5-formylamino-6-ribosylaminopyrimidin-4(3H)-one 5'-monophosphate deformylase as the second step in the archaeal riboflavin biosynthetic pathway.







