
In biochemistry, a kinase is an enzyme that catalyzes the transfer of phosphate groups from high-energy, phosphate-donating molecules to specific substrates. This process is known as phosphorylation, where the high-energy ATP molecule donates a phosphate group to the substrate molecule. This transesterification produces a phosphorylated substrate and ADP. Conversely, it is referred to as dephosphorylation when the phosphorylated substrate donates a phosphate group and ADP gains a phosphate group. These two processes, phosphorylation and dephosphorylation, occur four times during glycolysis.
In molecular biology, biosynthesis is a multi-step, enzyme-catalyzed process where substrates are converted into more complex products in living organisms. In biosynthesis, simple compounds are modified, converted into other compounds, or joined to form macromolecules. This process often consists of metabolic pathways. Some of these biosynthetic pathways are located within a single cellular organelle, while others involve enzymes that are located within multiple cellular organelles. Examples of these biosynthetic pathways include the production of lipid membrane components and nucleotides. Biosynthesis is usually synonymous with anabolism.
A heptose is a monosaccharide with seven carbon atoms.
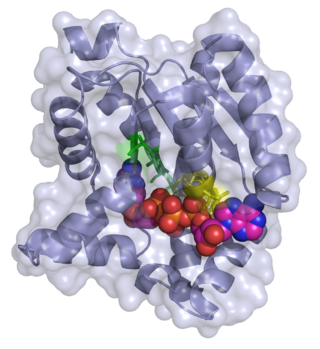
Adenylate kinase is a phosphotransferase enzyme that catalyzes the interconversion of the various adenosine phosphates. By constantly monitoring phosphate nucleotide levels inside the cell, ADK plays an important role in cellular energy homeostasis.

Nucleoside-diphosphate kinases are enzymes that catalyze the exchange of terminal phosphate between different nucleoside diphosphates (NDP) and triphosphates (NTP) in a reversible manner to produce nucleotide triphosphates. Many NDP serve as acceptor while NTP are donors of phosphate group. The general reaction via ping-pong mechanism is as follows: XDP + YTP ←→ XTP + YDP. NDPK activities maintain an equilibrium between the concentrations of different nucleoside triphosphates such as, for example, when guanosine triphosphate (GTP) produced in the citric acid (Krebs) cycle is converted to adenosine triphosphate (ATP). Other activities include cell proliferation, differentiation and development, signal transduction, G protein-coupled receptor, endocytosis, and gene expression.

Phosphoglycerate kinase is an enzyme that catalyzes the reversible transfer of a phosphate group from 1,3-bisphosphoglycerate (1,3-BPG) to ADP producing 3-phosphoglycerate (3-PG) and ATP :
Pantothenate kinase (EC 2.7.1.33, PanK; CoaA) is the first enzyme in the Coenzyme A (CoA) biosynthetic pathway. It phosphorylates pantothenate (vitamin B5) to form 4'-phosphopantothenate at the expense of a molecule of adenosine triphosphate (ATP). It is the rate-limiting step in the biosynthesis of CoA.
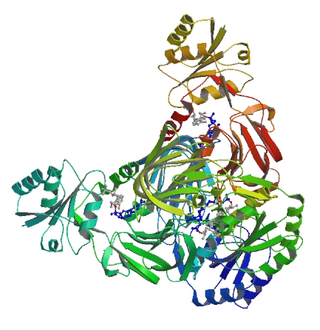
NAD+ kinase (EC 2.7.1.23, NADK) is an enzyme that converts nicotinamide adenine dinucleotide (NAD+) into NADP+ through phosphorylating the NAD+ coenzyme. NADP+ is an essential coenzyme that is reduced to NADPH primarily by the pentose phosphate pathway to provide reducing power in biosynthetic processes such as fatty acid biosynthesis and nucleotide synthesis. The structure of the NADK from the archaean Archaeoglobus fulgidus has been determined.
In enzymology, an ADP-L-glycero-D-manno-heptose 6-epimerase is an enzyme that catalyzes the chemical reaction
The Walker A and Walker B motifs are protein sequence motifs, known to have highly conserved three-dimensional structures. These were first reported in ATP-binding proteins by Walker and co-workers in 1982.
D-glycero-beta-D-manno-heptose-7-phosphate kinase is an enzyme with systematic name ATP:D-glycero-beta-D-manno-heptose 7-phosphate 1-phosphotransferase. This enzyme catalyses the following chemical reaction
D-beta-D-heptose 7-phosphate kinase/D-beta-D-heptose 1-phosphate adenylyltransferase may refer to:
RfaE (gene) may refer to:
D-glycero-beta-D-manno-heptose 1-phosphate adenylyltransferase is an enzyme with systematic name ATP:D-glycero-beta-D-manno-heptose 1-phosphate adenylyltransferase. This enzyme catalyses the following chemical reaction
D-glycero-alpha-D-manno-heptose 1-phosphate guanylyltransferase is an enzyme with systematic name GTP:D-glycero-alpha-D-manno-heptose 1-phosphate guanylyltransferase. This enzyme catalyses the following chemical reaction
D-glycero-β-D-manno-heptose 1,7-bisphosphate 7-phosphatase (EC 3.1.3.82) is an enzyme with systematic name D-glycero-β-D-manno-heptose 1,7-bisphosphate 7-phosphohydrolase. This enzyme catalyses the following chemical reaction
D-glycero-α-D-manno-heptose 1,7-bisphosphate 7-phosphatase (EC 3.1.3.83) is an enzyme with systematic name D-glycero-α-D-manno-heptose 1,7-bisphosphate 7-phosphohydrolase. This enzyme catalyses the following chemical reaction
D-sedoheptulose 7-phosphate isomerase is an enzyme with systematic name D-glycero-D-manno-heptose 7-phosphate aldose-ketose-isomerase. This enzyme catalyses the following chemical reaction





