Related Research Articles

Tryptase is the most abundant secretory granule-derived serine proteinase contained in mast cells and has been used as a marker for mast cell activation. Club cells contain tryptase, which is believed to be responsible for cleaving the hemagglutinin surface protein of influenza A virus, thereby activating it and causing the symptoms of flu.

Proteinase 3, also known as PRTN3, is an enzyme that in humans is encoded by the PRTN3 gene.

Endopeptidase Clp (EC 3.4.21.92, endopeptidase Ti, caseinolytic protease, protease Ti, ATP-dependent Clp protease, ClpP, Clp protease). This enzyme catalyses the following chemical reaction

Subtilisin is a protease initially obtained from Bacillus subtilis.
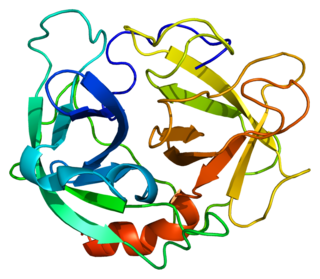
Neutrophil elastase is a serine proteinase in the same family as chymotrypsin and has broad substrate specificity. Neutrophil elastase is secreted by neutrophils during inflammation, and destroys bacteria and host tissue. It also localizes to neutrophil extracellular traps (NETs), via its high affinity for DNA, an unusual property for serine proteases.

Cathepsin G is a protein that in humans is encoded by the CTSG gene. It is one of the three serine proteases of the chymotrypsin family that are stored in the azurophil granules, and also a member of the peptidase S1 protein family. Cathepsin G plays an important role in eliminating intracellular pathogens and breaking down tissues at inflammatory sites, as well as in anti-inflammatory response.
In enzymology, a [acyl-carrier-protein] S-malonyltransferase is an enzyme that catalyzes the chemical reaction

Lon protease homolog, mitochondrial is a protease, an enzyme that in humans is encoded by the LONP1 gene.

Malate dehydrogenase, mitochondrial also known as malate dehydrogenase 2 is an enzyme that in humans is encoded by the MDH2 gene.

ATP-dependent Clp protease proteolytic subunit (ClpP) is an enzyme that in humans is encoded by the CLPP gene. This protein is an essential component to form the protein complex of Clp protease.

Morpheeins are proteins that can form two or more different homo-oligomers, but must come apart and change shape to convert between forms. The alternate shape may reassemble to a different oligomer. The shape of the subunit dictates which oligomer is formed. Each oligomer has a finite number of subunits (stoichiometry). Morpheeins can interconvert between forms under physiological conditions and can exist as an equilibrium of different oligomers. These oligomers are physiologically relevant and are not misfolded protein; this distinguishes morpheeins from prions and amyloid. The different oligomers have distinct functionality. Interconversion of morpheein forms can be a structural basis for allosteric regulation, an idea noted many years ago, and later revived. A mutation that shifts the normal equilibrium of morpheein forms can serve as the basis for a conformational disease. Features of morpheeins can be exploited for drug discovery. The dice image represents a morpheein equilibrium containing two different monomeric shapes that dictate assembly to a tetramer or a pentamer. The one protein that is established to function as a morpheein is porphobilinogen synthase, though there are suggestions throughout the literature that other proteins may function as morpheeins.

In molecular biology, the CLP protease family is a family of serine peptidases belong to the MEROPS peptidase family S14. ClpP is an ATP-dependent protease that cleaves a number of proteins, such as casein and albumin. It exists as a heterodimer of ATP-binding regulatory A and catalytic P subunits, both of which are required for effective levels of protease activity in the presence of ATP, although the P subunit alone does possess some catalytic activity.

In molecular biology, the Lon protease family is a family of enzymes that break peptide bonds in proteins resulting in smaller peptides or amino acids. They are found in archaea, bacteria and eukaryotes. Lon proteases are ATP-dependent serine peptidases belonging to the MEROPS peptidase family S16. In the eukaryotes the majority of the Lon proteases are located in the mitochondrial matrix. In yeast, the Lon protease PIM1 is located in the mitochondrial matrix. It is required for mitochondrial function, it is constitutively expressed but is increased after thermal stress, suggesting that PIM1 may play a role in the heat shock response. Lon proteases have two specific subfamilies: LonA and LonB, differentiated by the number of AAA+ domains found in the protein.

Glutamyl endopeptidase is an extracellular bacterial serine protease of the glutamyl endopeptidase I family that was initially isolated from the Staphylococcus aureus strain V8. The protease is, hence, commonly referred to as "V8 protease", or alternatively SspA from its corresponding gene.
Lysyl endopeptidase is an enzyme. This enzyme catalyses the following chemical reaction
Endopeptidase So is an enzyme. This enzyme catalyses the following chemical reaction

Peptidase Do is an enzyme. This enzyme catalyses the following chemical reaction
Pitrilysin is an enzyme. This enzyme catalyses the following chemical reaction:

ATP-dependent Clp protease ATP-binding subunit clpX-like, mitochondrial is an enzyme that in humans is encoded by the CLPX gene. This protein is a member of the family of AAA Proteins and is to form the protein complex of Clp protease.
Alfred Lewis Goldberg was an American cell biologist-biochemist and professor at Harvard University. His major discoveries have concerned the mechanisms and physiological importance of protein degradation in cells. Of wide impact have been his lab's demonstration that all cells contain a pathway for selectively eliminating misfolded proteins, his discoveries about the role of proteasomes in this process and of the enzyme systems catalyzing protein breakdown in bacteria, his elucidating the mechanisms for muscle atrophy and the role of proteasomes in antigen presentation to the immune system, and his introduction of proteasome inhibitors now widely used as research tools and in the treatment of blood cancers.
References
- ↑ Desautels M, Goldberg AL (October 1982). "Demonstration of an ATP-dependent, vanadate-sensitive endoprotease in the matrix of rat liver mitochondria". The Journal of Biological Chemistry. 257 (19): 11673–9. doi: 10.1016/S0021-9258(18)33815-8 . PMID 6749845.
- ↑ Larimore FS, Waxman L, Goldberg AL (April 1982). "Studies of the ATP-dependent proteolytic enzyme, protease La, from Escherichia coli". The Journal of Biological Chemistry. 257 (8): 4187–95. doi: 10.1016/S0021-9258(18)34704-5 . PMID 7040380.
- ↑ Chin DT, Goff SA, Webster T, Smith T, Goldberg AL (August 1988). "Sequence of the lon gene in Escherichia coli. A heat-shock gene which encodes the ATP-dependent protease La". The Journal of Biological Chemistry. 263 (24): 11718–28. doi: 10.1016/S0021-9258(18)37843-8 . PMID 3042779.