Related Research Articles

A protease is an enzyme that catalyzes proteolysis, breaking down proteins into smaller polypeptides or single amino acids, and spurring the formation of new protein products. They do this by cleaving the peptide bonds within proteins by hydrolysis, a reaction where water breaks bonds. Proteases are involved in many biological functions, including digestion of ingested proteins, protein catabolism, and cell signaling.

The hepatitis C virus (HCV) is a small, enveloped, positive-sense single-stranded RNA virus of the family Flaviviridae. The hepatitis C virus is the cause of hepatitis C and some cancers such as liver cancer and lymphomas in humans.
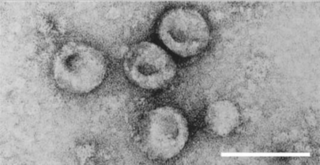
Pestivirus is a genus of viruses, in the family Flaviviridae. Viruses in the genus Pestivirus infect mammals, including members of the family Bovidae and the family Suidae. There are 11 species in this genus. Diseases associated with this genus include: hemorrhagic syndromes, abortion, and fatal mucosal disease.

TEV protease is a highly sequence-specific cysteine protease from Tobacco Etch Virus (TEV). It is a member of the PA clan of chymotrypsin-like proteases. Due to its high sequence specificity, TEV protease is frequently used for the controlled cleavage of fusion proteins in vitro and in vivo.
NS2-3 protease is an enzyme responsible for proteolytic cleavage between NS2 and NS3, which are non-structural proteins that form part of the HCV virus particle. NS3 protease of hepatitis C virus, on the other hand, is responsible for the cleavage of non-structural protein downstream. Both of these proteases are directly involved in HCV genome replication, that is, during the viral life-cycle that leads to virus multiplication in the host that has been infected by the virus.

Nonstructural protein 3 (NS3), also known as p-70, is a viral nonstructural protein that is 70 kDa cleavage product of the hepatitis C virus polyprotein. It acts as a serine protease. C-terminal two-thirds of the protein also acts as helicase and nucleoside triphosphatase. First (N-terminal) 180 aminoacids of NS3 has additional role as cofactor domains for NS2 protein.

Nonstructural protein 5A (NS5A) is a zinc-binding and proline-rich hydrophilic phosphoprotein that plays a key role in Hepatitis C virus RNA replication. It appears to be a dimeric form without trans-membrane helices.

Nonstructural protein 2 (NS2) is a viral protein found in the hepatitis C virus. It is also produced by influenza viruses, and is alternatively known as the nuclear export protein (NEP).

Ciluprevir was a drug used experimentally in the treatment of hepatitis C. It is manufactured by Boehringer Ingelheim and developed under the research code of BILN 2061. It was the first-in-class NS3/4A protease inhibitor to enter clinical development and tested in human. Ciluprevir is a potent competitive reversible inhibitor of NS3/4A protease from HCV genotype 1a (Ki = 0.3 nM) and 1b (Ki = 0.66 nM). It shows good selectivity for NS3 protease against representative serine and cysteine proteases, human leukocyte elastase and cathepsin B (IC50 > 30 μM).
Hepacivirus A, or Canine hepacivirus (CHV) or Equine hepacivirus (EHV), is a positive-sense single-stranded RNA virus of the genus Hepacivirus. It infects dogs and horses, and causes pulmonary infections in dogs. Unlike the related Hepatitis C virus, it is not known to cause hepatitis in either host.

Asunaprevir is an experimental drug candidate for the treatment of hepatitis C. It was undergoing development by Bristol-Myers Squibb and has completed Phase III clinical trials in 2013.
Carboxypeptidase D can refer to one of several enzymes. A family of serine carboxypeptidases includes is an enzyme. This enzyme has an optimal pH of 4.5-6.0, is inhibited by diisopropyl fluorophosphate, and catalyses the following chemical reaction
Flavivirin is an enzyme.
Pestivirus NS3 polyprotein peptidase is an enzyme. This enzyme catalyses the following chemical reaction

The 3C-like protease (3CLpro) or main protease (Mpro), formally known as C30 endopeptidase or 3-chymotrypsin-like protease, is the main protease found in coronaviruses. It cleaves the coronavirus polyprotein at eleven conserved sites. It is a cysteine protease and a member of the PA clan of proteases. It has a cysteine-histidine catalytic dyad at its active site and cleaves a Gln–(Ser/Ala/Gly) peptide bond.
HIV-2 retropepsin is an enzyme. This enzyme catalyses the following chemical reaction

The PA clan is the largest group of proteases with common ancestry as identified by structural homology. Members have a chymotrypsin-like fold and similar proteolysis mechanisms but can have identity of <10%. The clan contains both cysteine and serine proteases. PA clan proteases can be found in plants, animals, fungi, eubacteria, archaea and viruses.
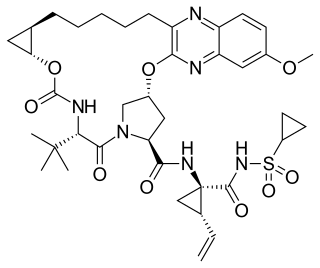
Grazoprevir is a drug approved for the treatment of hepatitis C. It was developed by Merck and completed Phase III trials, used in combination with the NS5A replication complex inhibitor elbasvir under the trade name Zepatier, either with or without ribavirin.

Glutamic proteases are a group of proteolytic enzymes containing a glutamic acid residue within the active site. This type of protease was first described in 2004 and became the sixth catalytic type of protease. Members of this group of protease had been previously assumed to be an aspartate protease, but structural determination showed it to belong to a novel protease family. The first structure of this group of protease was scytalidoglutamic peptidase, the active site of which contains a catalytic dyad, glutamic acid (E) and glutamine (Q), which give rise to the name eqolisin. This group of proteases are found primarily in pathogenic fungi affecting plant and human.
Entebbe bat virus is an infectious disease caused by a Flavivirus that is closely related to yellow fever.
References
- ↑ Kim JL, Morgenstern KA, Lin C, Fox T, Dwyer MD, Landro JA, Chambers SP, Markland W, Lepre CA, O'Malley ET, Harbeson SL, Rice CM, Murcko MA, Caron PR, Thomson JA (October 1996). "Crystal structure of the hepatitis C virus NS3 protease domain complexed with a synthetic NS4A cofactor peptide". Cell. 87 (2): 343–55. doi: 10.1016/s0092-8674(00)81351-3 . PMID 8861917.
- ↑ Rice, C.M.; Rawlings, N.D.; Woessner, J.F. (1998). "Hepatitis C virus polyprotein peptidase". In Barrett, A.J. (ed.). Handbook of Proteolytic Enzymes. London: Academic Press. pp. 272–277.