Related Research Articles

Chymosin or rennin is a protease found in rennet. It is an aspartic endopeptidase belonging to MEROPS A1 family. It is produced by newborn ruminant animals in the lining of the abomasum to curdle the milk they ingest, allowing a longer residence in the bowels and better absorption. It is widely used in the production of cheese.
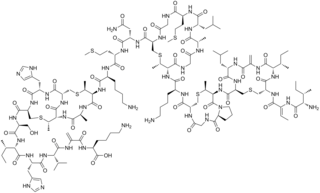
Nisin is a polycyclic antibacterial peptide produced by the bacterium Lactococcus lactis that is used as a food preservative. It has 34 amino acid residues, including the uncommon amino acids lanthionine (Lan), methyllanthionine (MeLan), didehydroalanine (Dha), and didehydroaminobutyric acid (Dhb). These unusual amino acids are introduced by posttranslational modification of the precursor peptide. In these reactions a ribosomally synthesized 57-mer is converted to the final peptide. The unsaturated amino acids originate from serine and threonine, and the enzyme-catalysed addition of cysteine residues to the didehydro amino acids result in the multiple (5) thioether bridges.
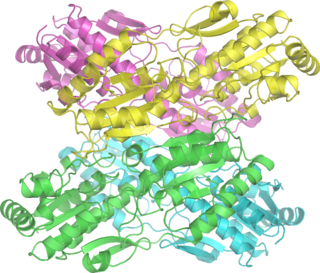
Phosphofructokinase-1 (PFK-1) is one of the most important regulatory enzymes of glycolysis. It is an allosteric enzyme made of 4 subunits and controlled by many activators and inhibitors. PFK-1 catalyzes the important "committed" step of glycolysis, the conversion of fructose 6-phosphate and ATP to fructose 1,6-bisphosphate and ADP. Glycolysis is the foundation for respiration, both anaerobic and aerobic. Because phosphofructokinase (PFK) catalyzes the ATP-dependent phosphorylation to convert fructose-6-phosphate into fructose 1,6-bisphosphate and ADP, it is one of the key regulatory steps of glycolysis. PFK is able to regulate glycolysis through allosteric inhibition, and in this way, the cell can increase or decrease the rate of glycolysis in response to the cell's energy requirements. For example, a high ratio of ATP to ADP will inhibit PFK and glycolysis. The key difference between the regulation of PFK in eukaryotes and prokaryotes is that in eukaryotes PFK is activated by fructose 2,6-bisphosphate. The purpose of fructose 2,6-bisphosphate is to supersede ATP inhibition, thus allowing eukaryotes to have greater sensitivity to regulation by hormones like glucagon and insulin.

Lactococcus is a genus of lactic acid bacteria that were formerly included in the genus Streptococcus Group N1. They are known as homofermenters meaning that they produce a single product, lactic acid in this case, as the major or only product of glucose fermentation. Their homofermentative character can be altered by adjusting environmental conditions such as pH, glucose concentration, and nutrient limitation. They are gram-positive, catalase-negative, non-motile cocci that are found singly, in pairs, or in chains. The genus contains strains known to grow at or below 7˚C.

Lactococcus lactis is a gram-positive bacterium used extensively in the production of buttermilk and cheese, but has also become famous as the first genetically modified organism to be used alive for the treatment of human disease. L. lactis cells are cocci that group in pairs and short chains, and, depending on growth conditions, appear ovoid with a typical length of 0.5 - 1.5 µm. L. lactis does not produce spores (nonsporulating) and are not motile (nonmotile). They have a homofermentative metabolism, meaning they produce lactic acid from sugars. They've also been reported to produce exclusive L-(+)-lactic acid. However, reported D-(−)-lactic acid can be produced when cultured at low pH. The capability to produce lactic acid is one of the reasons why L. lactis is one of the most important microorganisms in the dairy industry. Based on its history in food fermentation, L. lactis has generally recognized as safe (GRAS) status, with few case reports of it being an opportunistic pathogen.

Blue cheese is any of a wide range of cheeses made with the addition of cultures of edible molds, which create blue-green spots or veins through the cheese. Blue cheeses vary in taste from very mild to strong, and from slightly sweet to salty or sharp; in colour from pale to dark; and in consistency from liquid or very soft to firm or hard. They may have a distinctive smell, either from the mold or from various specially cultivated bacteria such as Brevibacterium linens.
Protease-activated receptors(PAR) are a subfamily of related G protein-coupled receptors that are activated by cleavage of part of their extracellular domain. They are highly expressed in platelets, and also on endothelial cells, fibroblasts, immune cells, myocytes, neurons, and tissues that line the gastrointestinal tract.

Nidovirales is an order of enveloped, positive-strand RNA viruses which infect vertebrates and invertebrates. Host organisms include mammals, birds, reptiles, amphibians, fish, arthropods, molluscs, and helminths. The order includes the families Coronaviridae, Arteriviridae, Roniviridae, and Mesoniviridae.

Cysteine proteases, also known as thiol proteases, are hydrolase enzymes that degrade proteins. These proteases share a common catalytic mechanism that involves a nucleophilic cysteine thiol in a catalytic triad or dyad.

Lactobacillales are an order of gram-positive, low-GC, acid-tolerant, generally nonsporulating, nonrespiring, either rod-shaped (bacilli) or spherical (cocci) bacteria that share common metabolic and physiological characteristics. These bacteria, usually found in decomposing plants and milk products, produce lactic acid as the major metabolic end product of carbohydrate fermentation, giving them the common name lactic acid bacteria (LAB).
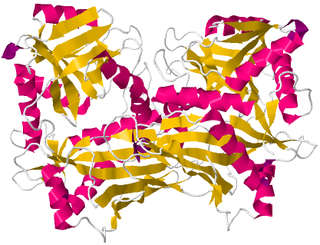
Branched-chain amino acid aminotransferase (BCAT), also known as branched-chain amino acid transaminase, is an aminotransferase enzyme (EC 2.6.1.42) which acts upon branched-chain amino acids (BCAAs). It is encoded by the BCAT2 gene in humans. The BCAT enzyme catalyzes the conversion of BCAAs and α-ketoglutarate into branched chain α-keto acids and glutamate.

In molecular biology, Proteinase K is a broad-spectrum serine protease. The enzyme was discovered in 1974 in extracts of the fungus Parengyodontium album. Proteinase K is able to digest hair (keratin), hence, the name "Proteinase K". The predominant site of cleavage is the peptide bond adjacent to the carboxyl group of aliphatic and aromatic amino acids with blocked alpha amino groups. It is commonly used for its broad specificity. This enzyme belongs to Peptidase family S8 (subtilisin). The molecular weight of Proteinase K is 28,900 daltons.
Κ-casein, or kappa casein, is a mammalian milk protein involved in several important physiological processes. Chymosin splits K-casein into an insoluble peptide and water-soluble glycomacropeptide (GMP). GMP is responsible for an increased efficiency of digestion, prevention of neonate hypersensitivity to ingested proteins, and inhibition of gastric pathogens. The human gene for κ-casein is CSN3.
Xaa-Pro dipeptidyl-peptidase (EC 3.4.14.11, X-prolyl dipeptidyl aminopeptidase, PepX, X-prolyl dipeptidyl peptidase is an enzyme. It catalyses the following chemical reaction

Zingibain, zingipain, or ginger protease is a cysteine protease enzyme found in ginger rhizomes. It catalyses the preferential cleavage of peptides with a proline residue at the P2 position. It has two distinct forms, ginger protease I (GP-I) and ginger protease II (GP-II).

Lancefield grouping is a system of classification that classifies catalase-negative Gram-positive cocci based on the carbohydrate composition of bacterial antigens found on their cell walls. The system, created by Rebecca Lancefield, was historically used to organize the various members of the family Streptococcaceae, which includes the genera Lactococcus and Streptococcus, but now is largely superfluous due to explosive growth in the number of streptococcal species identified since the 1970s. However, it has retained some clinical usefulness even after the taxonomic changes, and as of 2018, Lancefield designations are still often used to communicate medical microbiological test results.
Changestat is a continuous cultivation method that is used for acquiring quantitative data of a microorganism's metabolism at various environmental conditions within a single experiment. Every changestat always starts as a continuous cultivation experiment, but after reaching steady state, smooth and slow change of an environmental parameter is applied. Two most common changestat techniques are accelerostat (A-stat) and dilution rate stat (D-stat).
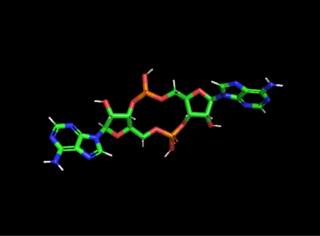
Cyclic di-AMP is a second messenger used in signal transduction in bacteria and archaea. It is present in many Gram-positive bacteria, some Gram-negative species, and archaea of the phylum euryarchaeota.
The Lactococcus lactis Phage r1t Holin Family is a family of putative pore-forming proteins that typically range in size between about 65 and 95 amino acyl residues (aas) in length, although a few r1t holins have been found to be significantly larger. Phage r1t holins exhibit between 2 and 4 transmembrane segments (TMSs), with the 4 TMS proteins resulting from an intragenic duplication of a 2 TMS region. A representative list of the proteins belonging to the r1t holin family can be found in the Transporter Classification Database.
The Bacterophase Dp-1 Holin Family is a family of proteins present in several Gram-positive bacteria and their phage. The genes coding for the lytic system of the pneumococcal phage, Dp-1, has been cloned and characterized. The holin of phage Dp-1 is 74 amino acyl residues (aas) long with two putative transmembrane segments (TMSs). The lytic enzyme of Dp-1 (Pal), an N-acetyl-muramoyl-L-alanine amidase, shows a modular organization similar to that described for the lytic enzymes of Streptococcus pneumoniae and its bacteriophage in which change in the order of the functional domains changes the enzyme specificity. A representative list of proteins belonging to the Dp-1 family can be found in the Transporter Classification Database.
References
- ↑ Visser S, Robben AJ, Slangen CJ (1991). "Specificity of a cell-envelope-located proteinase (PIII-type) from Lactococcus lactis subsp. cremoris AM1 in its action on bovine β-casein". Appl. Microbiol. Biotechnol. 35 (4): 477–483. doi:10.1007/bf00169753. PMID 1367552.
- ↑ Exterkate FA, Alting AC, Bruinenberg PG (November 1993). "Diversity of cell envelope proteinase specificity among strains of Lactococcus lactis and its relationship to charge characteristics of the substrate-binding region". Applied and Environmental Microbiology. 59 (11): 3640–7. PMC 182510 . PMID 8285671.
- ↑ Pritchard GG, Coolbear T (September 1993). "The physiology and biochemistry of the proteolytic system in lactic acid bacteria". FEMS Microbiology Reviews. 12 (1–3): 179–206. doi: 10.1111/j.1574-6976.1993.tb00018.x . PMID 8398214.