Related Research Articles

Enzymes are proteins that act as biological catalysts by accelerating chemical reactions. The molecules upon which enzymes may act are called substrates, and the enzyme converts the substrates into different molecules known as products. Almost all metabolic processes in the cell need enzyme catalysis in order to occur at rates fast enough to sustain life. Metabolic pathways depend upon enzymes to catalyze individual steps. The study of enzymes is called enzymology and the field of pseudoenzyme analysis recognizes that during evolution, some enzymes have lost the ability to carry out biological catalysis, which is often reflected in their amino acid sequences and unusual 'pseudocatalytic' properties.
In biochemistry, a ligase is an enzyme that can catalyze the joining (ligation) of two molecules by forming a new chemical bond. This is typically via hydrolysis of a small pendant chemical group on one of the molecules, typically resulting in the formation of new C-O, C-S, or C-N bonds. For example, DNA ligase can join two complementary fragments of nucleic acid by forming phosphodiester bonds, and repair single stranded breaks that arise in double stranded DNA during replication.
A restriction enzyme, restriction endonuclease, REase, ENase orrestrictase is an enzyme that cleaves DNA into fragments at or near specific recognition sites within molecules known as restriction sites. Restriction enzymes are one class of the broader endonuclease group of enzymes. Restriction enzymes are commonly classified into five types, which differ in their structure and whether they cut their DNA substrate at their recognition site, or if the recognition and cleavage sites are separate from one another. To cut DNA, all restriction enzymes make two incisions, once through each sugar-phosphate backbone of the DNA double helix.

The enzyme-linked immunosorbent assay (ELISA) is a commonly used analytical biochemistry assay, first described by Eva Engvall and Peter Perlmann in 1971. The assay uses a solid-phase type of enzyme immunoassay (EIA) to detect the presence of a ligand in a liquid sample using antibodies directed against the protein to be measured. ELISA has been used as a diagnostic tool in medicine, plant pathology, and biotechnology, as well as a quality control check in various industries.

In biology and biochemistry, the active site is the region of an enzyme where substrate molecules bind and undergo a chemical reaction. The active site consists of amino acid residues that form temporary bonds with the substrate, the binding site, and residues that catalyse a reaction of that substrate, the catalytic site. Although the active site occupies only ~10–20% of the volume of an enzyme, it is the most important part as it directly catalyzes the chemical reaction. It usually consists of three to four amino acids, while other amino acids within the protein are required to maintain the tertiary structure of the enzymes.

In biochemistry, Michaelis–Menten kinetics, named after Leonor Michaelis and Maud Menten, is the simplest case of enzyme kinetics, applied to enzyme-catalysed reactions of one substrate and one product. It takes the form of an equation describing the rate reaction rate to , the concentration of the substrate A. Its formula is given by the Michaelis–Menten equation:

Nicotinamide adenine dinucleotide (NAD) is a coenzyme central to metabolism. Found in all living cells, NAD is called a dinucleotide because it consists of two nucleotides joined through their phosphate groups. One nucleotide contains an adenine nucleobase and the other, nicotinamide. NAD exists in two forms: an oxidized and reduced form, abbreviated as NAD+ and NADH (H for hydrogen), respectively.

A cofactor is a non-protein chemical compound or metallic ion that is required for an enzyme's role as a catalyst. Cofactors can be considered "helper molecules" that assist in biochemical transformations. The rates at which these happen are characterized in an area of study called enzyme kinetics. Cofactors typically differ from ligands in that they often derive their function by remaining bound.

Digestive enzymes are a group of enzymes that break down polymeric macromolecules into their smaller building blocks, in order to facilitate their absorption into the cells of the body. Digestive enzymes are found in the digestive tracts of animals and in the tracts of carnivorous plants, where they aid in the digestion of food, as well as inside cells, especially in their lysosomes, where they function to maintain cellular survival. Digestive enzymes of diverse specificities are found in the saliva secreted by the salivary glands, in the secretions of cells lining the stomach, in the pancreatic juice secreted by pancreatic exocrine cells, and in the secretions of cells lining the small and large intestines.
BRENDA is an information system representing one of the most comprehensive enzyme repositories. It is an electronic resource that comprises molecular and biochemical information on enzymes that have been classified by the IUBMB. Every classified enzyme is characterized with respect to its catalyzed biochemical reaction. Kinetic properties of the corresponding reactants are described in detail. BRENDA contains enzyme-specific data manually extracted from primary scientific literature and additional data derived from automatic information retrieval methods such as text mining. It provides a web-based user interface that allows a convenient and sophisticated access to the data.

Enzyme kinetics is the study of the rates of enzyme-catalysed chemical reactions. In enzyme kinetics, the reaction rate is measured and the effects of varying the conditions of the reaction are investigated. Studying an enzyme's kinetics in this way can reveal the catalytic mechanism of this enzyme, its role in metabolism, how its activity is controlled, and how a drug or a modifier might affect the rate.

An enzyme inhibitor is a molecule that binds to an enzyme and blocks its activity. Enzymes are proteins that speed up chemical reactions necessary for life, in which substrate molecules are converted into products. An enzyme facilitates a specific chemical reaction by binding the substrate to its active site, a specialized area on the enzyme that accelerates the most difficult step of the reaction.
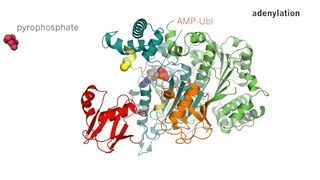
Enzyme catalysis is the increase in the rate of a process by a biological molecule, an "enzyme". Most enzymes are proteins, and most such processes are chemical reactions. Within the enzyme, generally catalysis occurs at a localized site, called the active site.
In chemistry, the term substrate is highly context-dependent. Broadly speaking, it can refer either to a chemical species being observed in a chemical reaction, or to a surface on which other chemical reactions or microscopy are performed.
Translocase is a general term for a protein that assists in moving another molecule, usually across a cell membrane. These enzymes catalyze the movement of ions or molecules across membranes or their separation within membranes. The reaction is designated as a transfer from “side 1” to “side 2” because the designations “in” and “out”, which had previously been used, can be ambiguous. Translocases are the most common secretion system in Gram positive bacteria.
Enzyme activators are molecules that bind to enzymes and increase their activity. They are the opposite of enzyme inhibitors. These molecules are often involved in the allosteric regulation of enzymes in the control of metabolism. An example of an enzyme activator working in this way is fructose 2,6-bisphosphate, which activates phosphofructokinase 1 and increases the rate of glycolysis in response to the hormone glucagon. In some cases, when a substrate binds to one catalytic subunit of an enzyme, this can trigger an increase in the substrate affinity as well as catalytic activity in the enzyme's other subunits, and thus the substrate acts as an activator.

A diffusion-limited enzyme catalyses a reaction so efficiently that the rate limiting step is that of substrate diffusion into the active site, or product diffusion out. This is also known as kinetic perfection or catalytic perfection. Since the rate of catalysis of such enzymes is set by the diffusion-controlled reaction, it therefore represents an intrinsic, physical constraint on evolution. Diffusion limited perfect enzymes are very rare. Most enzymes catalyse their reactions to a rate that is 1,000-10,000 times slower than this limit. This is due to both the chemical limitations of difficult reactions, and the evolutionary limitations that such high reaction rates do not confer any extra fitness.
PRIAM enzyme-specific profiles is a method for the automatic detection of likely enzymes in protein sequences. PRIAM uses position-specific scoring matrices automatically generated for each enzyme entry.
IntEnz contains data on enzymes organized by enzyme EC number and is the official version of the Enzyme Nomenclature system developed by the International Union of Biochemistry and Molecular Biology.
Enzyme promiscuity is the ability of an enzyme to catalyse a fortuitous side reaction in addition to its main reaction. Although enzymes are remarkably specific catalysts, they can often perform side reactions in addition to their main, native catalytic activity. These promiscuous activities are usually slow relative to the main activity and are under neutral selection. Despite ordinarily being physiologically irrelevant, under new selective pressures these activities may confer a fitness benefit therefore prompting the evolution of the formerly promiscuous activity to become the new main activity. An example of this is the atrazine chlorohydrolase from Pseudomonas sp. ADP that evolved from melamine deaminase, which has very small promiscuous activity toward atrazine, a man-made chemical.
References
- ↑ Gaucher GM, Stevenson KJ (1976). Thermomycolin. Methods in Enzymology. Vol. 45. pp. 415–33. doi:10.1016/s0076-6879(76)45037-1. PMID 1012007.