Related Research Articles
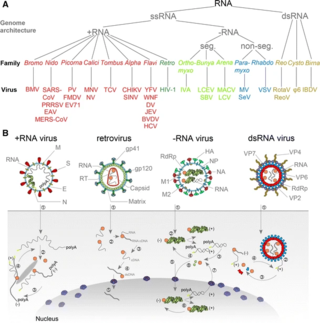
An RNA virus is a virus—other than a retrovirus—that has ribonucleic acid (RNA) as its genetic material. The nucleic acid is usually single-stranded RNA (ssRNA) but it may be double-stranded (dsRNA). Notable human diseases caused by RNA viruses include the common cold, influenza, SARS, MERS, Covid-19, Dengue Virus, hepatitis C, hepatitis E, West Nile fever, Ebola virus disease, rabies, polio, mumps, and measles.

A protease is an enzyme that catalyzes proteolysis, breaking down proteins into smaller polypeptides or single amino acids, and spurring the formation of new protein products. They do this by cleaving the peptide bonds within proteins by hydrolysis, a reaction where water breaks bonds. Proteases are involved in many biological functions, including digestion of ingested proteins, protein catabolism, and cell signaling.

Brome mosaic virus (BMV) is a small, positive-stranded, icosahedral RNA plant virus belonging to the genus Bromovirus, family Bromoviridae, in the Alphavirus-like superfamily.

A catalytic triad is a set of three coordinated amino acids that can be found in the active site of some enzymes. Catalytic triads are most commonly found in hydrolase and transferase enzymes. An acid-base-nucleophile triad is a common motif for generating a nucleophilic residue for covalent catalysis. The residues form a charge-relay network to polarise and activate the nucleophile, which attacks the substrate, forming a covalent intermediate which is then hydrolysed to release the product and regenerate free enzyme. The nucleophile is most commonly a serine or cysteine amino acid, but occasionally threonine or even selenocysteine. The 3D structure of the enzyme brings together the triad residues in a precise orientation, even though they may be far apart in the sequence.
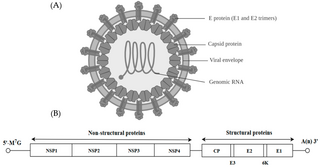
Alphavirus is a genus of RNA viruses, the sole genus in the Togaviridae family. Alphaviruses belong to group IV of the Baltimore classification of viruses, with a positive-sense, single-stranded RNA genome. There are 32 alphaviruses, which infect various vertebrates such as humans, rodents, fish, birds, and larger mammals such as horses, as well as invertebrates. Alphaviruses that could infect both vertebrates and arthropods are referred dual-host alphaviruses, while insect-specific alphaviruses such as Eilat virus and Yada yada virus are restricted to their competent arthropod vector. Transmission between species and individuals occurs mainly via mosquitoes, making the alphaviruses a member of the collection of arboviruses – or arthropod-borne viruses. Alphavirus particles are enveloped, have a 70 nm diameter, tend to be spherical, and have a 40 nm isometric nucleocapsid.

Cysteine proteases, also known as thiol proteases, are hydrolase enzymes that degrade proteins. These proteases share a common catalytic mechanism that involves a nucleophilic cysteine thiol in a catalytic triad or dyad.

Heparan sulfate (HS) is a linear polysaccharide found in all animal tissues. It occurs as a proteoglycan in which two or three HS chains are attached in close proximity to cell surface or extracellular matrix proteins. In this form, HS binds to a variety of protein ligands, including Wnt, and regulates a wide range of biological activities, including developmental processes, angiogenesis, blood coagulation, abolishing detachment activity by GrB, and tumour metastasis. HS has also been shown to serve as cellular receptor for a number of viruses, including the respiratory syncytial virus. One study suggests that cellular heparan sulfate has a role in SARS-CoV-2 Infection, particularly when the virus attaches with ACE2.

In molecular biology, Proteinase K is a broad-spectrum serine protease. The enzyme was discovered in 1974 in extracts of the fungus Parengyodontium album. Proteinase K is able to digest hair (keratin), hence, the name "Proteinase K". The predominant site of cleavage is the peptide bond adjacent to the carboxyl group of aliphatic and aromatic amino acids with blocked alpha amino groups. It is commonly used for its broad specificity. This enzyme belongs to Peptidase family S8 (subtilisin). The molecular weight of Proteinase K is 28,900 daltons.

Ubiquitin-like modifier-activating enzyme 7 is a protein that in humans is encoded by the UBA7 gene.
Mayaro virus disease is a mosquito-borne zoonotic pathogen endemic to certain humid forests of tropical South America. Infection with Mayaro virus causes an acute, self-limited dengue-like illness of 3–5 days' duration. The causative virus, abbreviated MAYV, is in the family Togaviridae, and genus Alphavirus. It is closely related to other alphaviruses that produce a dengue-like illness accompanied by long-lasting arthralgia. It is only known to circulate in tropical South America.
In molecular biology, the Bowman–Birk protease inhibitor family of proteins consists of eukaryotic proteinase inhibitors, belonging to MEROPS inhibitor family I12, clan IF. They mainly inhibit serine peptidases of the S1 family, but also inhibit S3 peptidases.
Oryzin is an enzyme. This enzyme catalyses the following chemical reaction
Streptogrisin A is an enzyme. This enzyme catalyses the following chemical reaction
Caricain is an enzyme. This enzyme catalyses the following chemical reaction: Hydrolysis of proteins with broad specificity for peptide bonds, similar to those of papain and chymopapain
Adenain is an enzyme. This enzyme catalyses the following chemical reaction
Saccharopepsin is an enzyme. This enzyme catalyses the following chemical reaction
Bacillolysin is an enzyme. This enzyme catalyses the following chemical reaction
Mitochondrial processing peptidase is an enzyme complex found in mitochondria which cleaves signal sequences from mitochondrial proteins. In humans this complex is composed of two subunits encoded by the genes PMPCA, and PMPCB. The enzyme is also known as. This enzyme catalyses the following chemical reaction

The PA clan is the largest group of proteases with common ancestry as identified by structural homology. Members have a chymotrypsin-like fold and similar proteolysis mechanisms but can have identity of <10%. The clan contains both cysteine and serine proteases. PA clan proteases can be found in plants, animals, fungi, eubacteria, archaea and viruses.

Glutamic proteases are a group of proteolytic enzymes containing a glutamic acid residue within the active site. This type of protease was first described in 2004 and became the sixth catalytic type of protease. Members of this group of protease had been previously assumed to be an aspartate protease, but structural determination showed it to belong to a novel protease family. The first structure of this group of protease was scytalidoglutamic peptidase, the active site of which contains a catalytic dyad, glutamic acid (E) and glutamine (Q), which give rise to the name eqolisin. This group of proteases are found primarily in pathogenic fungi affecting plant and human.
References
- ↑ Kräusslich HG, Wimmer E (1988). "Viral proteinases". Annual Review of Biochemistry. 57: 701–54. doi:10.1146/annurev.bi.57.070188.003413. PMID 3052288.
- ↑ Strauss EG, De Groot RJ, Levinson R, Strauss JH (December 1992). "Identification of the active site residues in the nsP2 proteinase of Sindbis virus". Virology. 191 (2): 932–40. doi:10.1016/0042-6822(92)90268-t. PMC 7131396 . PMID 1448929.
- ↑ Tong L, Wengler G, Rossmann MG (March 1993). "Refined structure of Sindbis virus core protein and comparison with other chymotrypsin-like serine proteinase structures". Journal of Molecular Biology. 230 (1): 228–47. doi:10.1006/jmbi.1993.1139. PMID 8450538.