Peptidoglycan or murein is a unique large macromolecule, a polysaccharide, consisting of sugars and amino acids that forms a mesh-like layer (sacculus) that surrounds the bacterial cytoplasmic membrane. The sugar component consists of alternating residues of β-(1,4) linked N-acetylglucosamine (NAG) and N-acetylmuramic acid (NAM). Attached to the N-acetylmuramic acid is an oligopeptide chain made of three to five amino acids. The peptide chain can be cross-linked to the peptide chain of another strand forming the 3D mesh-like layer. Peptidoglycan serves a structural role in the bacterial cell wall, giving structural strength, as well as counteracting the osmotic pressure of the cytoplasm. This repetitive linking results in a dense peptidoglycan layer which is critical for maintaining cell form and withstanding high osmotic pressures, and it is regularly replaced by peptidoglycan production. Peptidoglycan hydrolysis and synthesis are two processes that must occur in order for cells to grow and multiply, a technique carried out in three stages: clipping of current material, insertion of new material, and re-crosslinking of existing material to new material.
Phosphatidic acids are anionic phospholipids important to cell signaling and direct activation of lipid-gated ion channels. Hydrolysis of phosphatidic acid gives rise to one molecule each of glycerol and phosphoric acid and two molecules of fatty acids. They constitute about 0.25% of phospholipids in the bilayer.
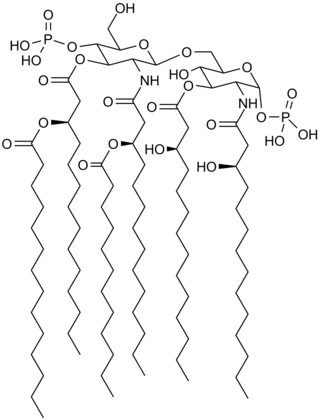
Lipid A is a lipid component of an endotoxin held responsible for the toxicity of gram-negative bacteria. It is the innermost of the three regions of the lipopolysaccharide (LPS), also called endotoxin molecule, and its hydrophobic nature allows it to anchor the LPS to the outer membrane. While its toxic effects can be damaging, the sensing of lipid A by the immune system may also be critical for the onset of immune responses to gram-negative infection, and for the subsequent successful fight against the infection.

The bacterial outer membrane is found in gram-negative bacteria. Gram-negative bacteria form two lipid bilayers in their cell envelopes - an inner membrane (IM) that encapsulates the cytoplasm, and an outer membrane (OM) that encapsulates the periplasm.

Sphingomyelin phosphodiesterase is a hydrolase enzyme that is involved in sphingolipid metabolism reactions. SMase is a member of the DNase I superfamily of enzymes and is responsible for breaking sphingomyelin (SM) down into phosphocholine and ceramide. The activation of SMase has been suggested as a major route for the production of ceramide in response to cellular stresses.

The enzyme UDP-glucose 4-epimerase, also known as UDP-galactose 4-epimerase or GALE, is a homodimeric epimerase found in bacterial, fungal, plant, and mammalian cells. This enzyme performs the final step in the Leloir pathway of galactose metabolism, catalyzing the reversible conversion of UDP-galactose to UDP-glucose. GALE tightly binds nicotinamide adenine dinucleotide (NAD+), a co-factor required for catalytic activity.

The enzyme phosphatidate phosphatase (PAP, EC 3.1.3.4) is a key regulatory enzyme in lipid metabolism, catalyzing the conversion of phosphatidate to diacylglycerol:
In enzymology, an acyl-[acyl-carrier-protein]-UDP-N-acetylglucosamine O-acyltransferase is an enzyme that catalyzes the chemical reaction
In enzymology, a lipid-A-disaccharide synthase is an enzyme that catalyzes the chemical reaction
In enzymology, a CDP-diacylglycerol—serine O-phosphatidyltransferase is an enzyme that catalyzes the chemical reaction

Saccharolipids are chemical compounds containing fatty acids linked directly to a sugar backbone, forming structures that are compatible with membrane bilayers. In the saccharolipids, a monosaccharide substitutes for the glycerol backbone present in glycerolipids and glycerophospholipids. The most familiar saccharolipids are the acylated glucosamine precursors of the lipid A component of the lipopolysaccharides in Gram-negative bacteria. Typical lipid A molecules are disaccharides of glucosamine, which are derivatized with as many as seven fatty-acyl chains. The minimal lipopolysaccharide required for growth in Escherichia coli is Kdo2-Lipid A, a hexa-acylated disaccharide of glucosamine (LipidA) that is glycosylated with two 3-deoxy-D-manno-octulosonic acid (Kdo) residues.
In molecular biology, the lipopolysaccharide kinase (Kdo/WaaP) family is a family of lipopolysaccharide kinases that includes lipopolysaccharide core heptose(I) kinase rfaP. Lipopolysaccharide core heptose(I) kinase rfaP is required for the addition of phosphate to O-4 of the first heptose residue of the lipopolysaccharide (LPS) inner core region. It has previously been shown that it is necessary for resistance to hydrophobic and polycationic antimicrobials in E. coli and that it is required for virulence in invasive strains of Salmonella enterica. The family also includes 3-deoxy-D-manno-octulosonic acid kinase from Haemophilus influenzae, which phosphorylates Kdo-lipid IV(A), a lipopolysaccharide precursor, and is involved in virulence.
UDP-glucuronic acid dehydrogenase (UDP-4-keto-hexauronic acid decarboxylating) (EC 1.1.1.305, UDP-GlcUA decarboxylase, ArnADH) is an enzyme with systematic name UDP-glucuronate:NAD+ oxidoreductase (decarboxylating). This enzyme catalyses the following chemical reaction
UDP-4-amino-4-deoxy-L-arabinose formyltransferase is an enzyme with systematic name 10-formyltetrahydrofolate:UDP-4-amino-4-deoxy-beta-L-arabinose N-formyltransferase. This enzyme catalyses the following chemical reaction
UDP-3-O-(3-hydroxymyristoyl)glucosamine N-acyltransferase is an enzyme with systematic name (3R)-3-hydroxymyristoyl-(acyl-carrier protein):UDP-3-O-( -3-hydroxymyristoyl)-alpha-D-glucosamine N-acetyltransferase. This enzyme catalyses the following chemical reaction
Lipid IVA 4-amino-4-deoxy-L-arabinosyltransferase is an enzyme with systematic name 4-amino-4-deoxy-alpha-L-arabinopyranosyl ditrans, octacis-undecaprenyl phosphate:lipid IVA 4-amino-4-deoxy-L-arabinopyranosyltransferase. This enzyme catalyses the following chemical reaction
Lipid IVA 3-deoxy-D-manno-octulosonic acid transferase is an enzyme with systematic name CMP-3-deoxy-D-manno-oct-2-ulosonate:lipid IVA 3-deoxy-D-manno-oct-2-ulosonate transferase. This enzyme catalyses the following chemical reaction
3-deoxy-D-manno-octulosonic acid kinase is an enzyme with systematic name ATP:(KDO)-lipid IVA 3-deoxy-alpha-D-manno-oct-2-ulopyranose 4-phosphotransferase. This enzyme catalyses the following chemical reaction
Undecaprenyl-phosphate 4-deoxy-4-formamido-L-arabinose transferase is an enzyme with systematic name UDP-4-amino-4-deoxy-alpha-L-arabinose:ditrans,octacis-undecaprenyl phosphate 4-amino-4-deoxy-alpha-L-arabinosyltransferase. This enzyme catalyses the following chemical reaction
UDP-2,3-diacylglucosamine diphosphatase (EC 3.6.1.54, UDP-2,3-diacylglucosamine hydrolase, UDP-2,3-diacylglucosamine pyrophosphatase, ybbF (gene), lpxH (gene)) is an enzyme with systematic name UDP-2,3-bis((3R)-3-hydroxymyristoyl)-alpha-D-glucosamine 2,3-bis((3R)-3-hydroxymyristoyl)-beta-D-glucosaminyl 1-phosphate phosphohydrolase. This enzyme catalyses the following chemical reaction





