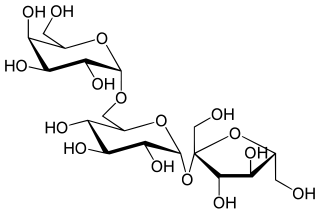
Raffinose is a trisaccharide composed of galactose, glucose, and fructose. It can be found in beans, cabbage, brussels sprouts, broccoli, asparagus, other vegetables, and whole grains. Raffinose can be hydrolyzed to D-galactose and sucrose by the enzyme α-galactosidase (α-GAL), an enzyme which in the lumen of the human digestive tract is only produced by bacteria in the large intestine. α-GAL also hydrolyzes other α-galactosides such as stachyose, verbascose, and galactinol, if present. The enzyme does not cleave β-linked galactose, as in lactose.
Glycogenin is an enzyme involved in converting glucose to glycogen. It acts as a primer, by polymerizing the first few glucose molecules, after which other enzymes take over. It is a homodimer of 37-kDa subunits and is classified as a glycosyltransferase.
In enzymology, a sequoyitol dehydrogenase (EC 1.1.1.143) is an enzyme that catalyzes the chemical reaction
In enzymology, a D-pinitol dehydrogenase (EC 1.1.1.142) is an enzyme that catalyzes the chemical reaction
In enzymology, a cholesterol 7alpha-monooxygenase (EC 1.14.13.17) is an enzyme that catalyzes the chemical reaction
In enzymology, an inositol-3-phosphate synthase is an enzyme that catalyzes the chemical reaction
The enzyme phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase (EC 3.1.3.67) catalyzes the chemical reaction
The enzyme phosphatidylinositol-3,4-bisphosphate 4-phosphatase (EC 3.1.3.66) that catalyzes the reaction
In enzymology, a N-acetylglucosaminylphosphatidylinositol deacetylase (EC 3.5.1.89) is an enzyme that catalyzes the chemical reaction
In enzymology, a chitobiosyldiphosphodolichol beta-mannosyltransferase is an enzyme that catalyzes the chemical reaction
In enzymology, a dolichyl-phosphate beta-D-mannosyltransferase is an enzyme that catalyzes the chemical reaction
In enzymology, a dolichyl-phosphate-mannose-protein mannosyltransferase is an enzyme that catalyzes the chemical reaction
In enzymology, a fucosylgalactoside 3-alpha-galactosyltransferase is an enzyme that catalyzes the chemical reaction
In enzymology, a galactinol-sucrose galactosyltransferase is an enzyme that catalyzes the chemical reaction
In enzymology, an inositol 3-alpha-galactosyltransferase is an enzyme that catalyzes the chemical reaction
In enzymology, a lactosylceramide beta-1,3-galactosyltransferase is an enzyme that catalyzes the chemical reaction
In enzymology, a monogalactosyldiacylglycerol synthase is an enzyme that catalyzes the chemical reaction

In enzymology, a CDP-diacylglycerol—inositol 3-phosphatidyltransferase is an enzyme that catalyzes the chemical reaction
Beta-1,4-galactosyltransferase 1 is an enzyme that in humans is encoded by the B4GALT1 gene.
A ureohydrolase is a type of hydrolase enzyme. The ureohydrolase superfamily includes arginase, agmatinase, formiminoglutamase and proclavaminate amidinohydrolase. These enzymes share a 3-layer alpha-beta-alpha structure, and play important roles in arginine/agmatine metabolism, the urea cycle, histidine degradation, and other pathways.