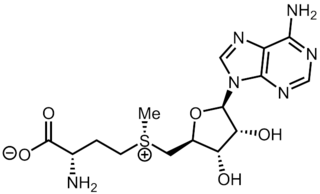
S-Adenosyl methionine (SAM), also known under the commercial names of SAMe, SAM-e, or AdoMet, is a common cosubstrate involved in methyl group transfers, transsulfuration, and aminopropylation. Although these anabolic reactions occur throughout the body, most SAM is produced and consumed in the liver. More than 40 methyl transfers from SAM are known, to various substrates such as nucleic acids, proteins, lipids and secondary metabolites. It is made from adenosine triphosphate (ATP) and methionine by methionine adenosyltransferase. SAM was first discovered by Giulio Cantoni in 1952.

Spermidine synthase is an enzyme that catalyzes the transfer of the propylamine group from S-adenosylmethioninamine to putrescine in the biosynthesis of spermidine. The systematic name is S-adenosyl 3-(methylthio)propylamine:putrescine 3-aminopropyltransferase and it belongs to the group of aminopropyl transferases. It does not need any cofactors. Most spermidine synthases exist in solution as dimers.

Methyltransferases are a large group of enzymes that all methylate their substrates but can be split into several subclasses based on their structural features. The most common class of methyltransferases is class I, all of which contain a Rossmann fold for binding S-Adenosyl methionine (SAM). Class II methyltransferases contain a SET domain, which are exemplified by SET domain histone methyltransferases, and class III methyltransferases, which are membrane associated. Methyltransferases can also be grouped as different types utilizing different substrates in methyl transfer reactions. These types include protein methyltransferases, DNA/RNA methyltransferases, natural product methyltransferases, and non-SAM dependent methyltransferases. SAM is the classical methyl donor for methyltransferases, however, examples of other methyl donors are seen in nature. The general mechanism for methyl transfer is a SN2-like nucleophilic attack where the methionine sulfur serves as the leaving group and the methyl group attached to it acts as the electrophile that transfers the methyl group to the enzyme substrate. SAM is converted to S-Adenosyl homocysteine (SAH) during this process. The breaking of the SAM-methyl bond and the formation of the substrate-methyl bond happen nearly simultaneously. These enzymatic reactions are found in many pathways and are implicated in genetic diseases, cancer, and metabolic diseases. Another type of methyl transfer is the radical S-Adenosyl methionine (SAM) which is the methylation of unactivated carbon atoms in primary metabolites, proteins, lipids, and RNA.

Ribose 5-phosphate (R5P) is both a product and an intermediate of the pentose phosphate pathway. The last step of the oxidative reactions in the pentose phosphate pathway is the production of ribulose 5-phosphate. Depending on the body's state, ribulose 5-phosphate can reversibly isomerize to ribose 5-phosphate. Ribulose 5-phosphate can alternatively undergo a series of isomerizations as well as transaldolations and transketolations that result in the production of other pentose phosphates as well as fructose 6-phosphate and glyceraldehyde 3-phosphate.
The enzyme adenosylmethionine decarboxylase catalyzes the conversion of S-adenosyl methionine to S-adenosylmethioninamine. Polyamines such as spermidine and spermine are essential for cellular growth under most conditions, being implicated in many cellular processes including DNA, RNA and protein synthesis. S-adenosylmethionine decarboxylase (AdoMetDC) plays an essential regulatory role in the polyamine biosynthetic pathway by generating the n-propylamine residue required for the synthesis of spermidine and spermine from putrescein. Unlike many amino acid decarboxylases AdoMetDC uses a covalently bound pyruvate residue as a cofactor rather than the more common pyridoxal 5'-phosphate. These proteins can be divided into two main groups which show little sequence similarity either to each other, or to other pyruvoyl-dependent amino acid decarboxylases: class I enzymes found in bacteria and archaea, and class II enzymes found in eukaryotes. In both groups the active enzyme is generated by the post-translational autocatalytic cleavage of a precursor protein. This cleavage generates the pyruvate precursor from an internal serine residue and results in the formation of two non-identical subunits termed alpha and beta which form the active enzyme.

ADP-ribosylation is the addition of one or more ADP-ribose moieties to a protein. It is a reversible post-translational modification that is involved in many cellular processes, including cell signaling, DNA repair, gene regulation and apoptosis. Improper ADP-ribosylation has been implicated in some forms of cancer. It is also the basis for the toxicity of bacterial compounds such as cholera toxin, diphtheria toxin, and others.
In enzymology, a rRNA (adenine-N6-)-methyltransferase (EC 2.1.1.48) is an enzyme that catalyzes the chemical reaction
In enzymology, a tRNA (adenine-N1-)-methyltransferase (EC 2.1.1.36) is an enzyme that catalyzes the chemical reaction
In enzymology, a tRNA (adenine-N6-)-methyltransferase (EC 2.1.1.55) is an enzyme that catalyzes the chemical reaction
In enzymology, a tRNA (guanine-N2-)-methyltransferase (EC 2.1.1.32) is an enzyme that catalyzes the chemical reaction
In enzymology, a preQ1 synthase (EC 1.7.1.13) is an enzyme that catalyzes the chemical reaction

The enzyme S-ribosylhomocysteine lyase catalyzes the reaction
In enzymology, a NAD(P)+-protein-arginine ADP-ribosyltransferase (EC 2.4.2.31) is an enzyme that catalyzes the chemical reaction using nicotinamide adenine dinucleotide
In enzymology, a S-methyl-5'-thioadenosine phosphorylase is an enzyme that catalyzes the chemical reaction

S-Adenosylmethionine synthetase, also known as methionine adenosyltransferase (MAT), is an enzyme that creates S-adenosylmethionine by reacting methionine and ATP.
Radical SAMenzymes is a superfamily of enzymes that use a [4Fe-4S]+ cluster to reductively cleave S-adenosyl-L-methionine (SAM) to generate a radical, usually a 5′-deoxyadenosyl radical (5'-dAdo), as a critical intermediate. These enzymes utilize this radical intermediate to perform diverse transformations, often to functionalize unactivated C-H bonds. Radical SAM enzymes are involved in cofactor biosynthesis, enzyme activation, peptide modification, post-transcriptional and post-translational modifications, metalloprotein cluster formation, tRNA modification, lipid metabolism, biosynthesis of antibiotics and natural products etc. The vast majority of known radical SAM enzymes belong to the radical SAM superfamily, and have a cysteine-rich motif that matches or resembles CxxxCxxC. Radical SAM enzymes comprise the largest superfamily of metal-containing enzymes.
23S rRNA (adenine2503-C2)-methyltransferase (EC 2.1.1.192, RlmN, YfgB, Cfr) is an enzyme with systematic name S-adenosyl-L-methionine:23S rRNA (adenine2503-C2)-methyltransferase. This enzyme catalyses the following chemical reaction
tRNA (cytidine56-2'-O)-methyltransferase is an enzyme with systematic name S-adenosyl-L-methionine:tRNA (cytidine56-2'-O)-methyltransferase. This enzyme catalyses the following chemical reaction
TRNA (guanine10-N2)-dimethyltransferase (EC 2.1.1.213, PAB1283, N(2),N(2)-dimethylguanosine tRNA methyltransferase, Trm-G10, PabTrm-G10, PabTrm-m2 2G10 enzyme) is an enzyme with systematic name S-adenosyl-L-methionine:tRNA (guanine10-N2)-dimethyltransferase. This enzyme catalyses the following chemical reaction
23S rRNA (adenine2503-C8)-methyltransferase (EC 2.1.1.224, Cfr (gene)) is an enzyme with systematic name S-adenosyl-L-methionine:23S rRNA (adenine2503-C8)-methyltransferase. This enzyme catalyses the following chemical reaction










