A salvage pathway is a pathway in which a biological product is produced from intermediates in the degradative pathway of its own or a similar substance. The term often refers to nucleotide salvage in particular, in which nucleotides are synthesized from intermediates in their degradative pathway.
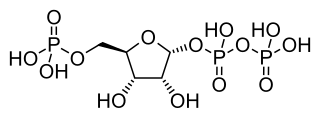
Phosphoribosyl pyrophosphate (PRPP) is a pentose phosphate. It is a biochemical intermediate in the formation of purine nucleotides via inosine-5-monophosphate, as well as in pyrimidine nucleotide formation. Hence it is a building block for DNA and RNA. The vitamins thiamine and cobalamin, and the amino acid tryptophan also contain fragments derived from PRPP. It is formed from ribose 5-phosphate (R5P) by the enzyme ribose-phosphate diphosphokinase:

Nucleic acid metabolism is a collective term that refers to the variety of chemical reactions by which nucleic acids are either synthesized or degraded. Nucleic acids are polymers made up of a variety of monomers called nucleotides. Nucleotide synthesis is an anabolic mechanism generally involving the chemical reaction of phosphate, pentose sugar, and a nitrogenous base. Degradation of nucleic acids is a catabolic reaction and the resulting parts of the nucleotides or nucleobases can be salvaged to recreate new nucleotides. Both synthesis and degradation reactions require multiple enzymes to facilitate the event. Defects or deficiencies in these enzymes can lead to a variety of diseases.

In enzymology, an anthranilate phosphoribosyltransferase is an enzyme that catalyzes the chemical reaction

In enzymology, an ATP phosphoribosyltransferase is an enzyme that catalyzes the chemical reaction
In enzymology, a dioxotetrahydropyrimidine phosphoribosyltransferase is an enzyme that catalyzes the chemical reaction

In enzymology, a nicotinate-nucleotide-dimethylbenzimidazole phosphoribosyltransferase is an enzyme that catalyzes the chemical reaction

In enzymology, a nicotinate-nucleotide diphosphorylase (carboxylating) (EC 2.4.2.19) is an enzyme that catalyzes the chemical reaction

In enzymology, a nicotinate phosphoribosyltransferase (EC 6.3.4.21) is an enzyme that catalyzes the chemical reaction
In enzymology, a xanthine phosphoribosyltransferase is an enzyme that catalyzes the chemical reaction
In enzymology, a ribose 1,5-bisphosphate phosphokinase is an enzyme that catalyzes the chemical reaction
Bisphosphate may refer to:
Rhamnopyranosyl-N-acetylglucosaminyl-diphospho-decaprenol beta-1,3/1,4-galactofuranosyltransferase is an enzyme with systematic name UDP-alpha-D-galactofuranose:alpha-L-rhamnopyranosyl-(1->3)-N-acetyl-alpha-D-glucosaminyl-diphospho-trans,octacis-decaprenol 3-beta/4-beta-galactofuranosyltransferase. This enzyme catalyses the following chemical reaction
Galactofuranosylgalactofuranosylrhamnosyl-N-acetylglucosaminyl-diphospho-decaprenol beta-1,5/1,6-galactofuranosyltransferase is an enzyme with systematic name UDP-alpha-D-galactofuranose:beta-D-galactofuranosyl-(1->5)-beta-D-galactofuranosyl-(1->4)-alpha-L-rhamnopyranosyl-(1->3)-N-acetyl-alpha-D-glucosaminyl-diphospho-trans,octacis-decaprenol 4-beta/5-beta-D-galactofuranosyltransferase. This enzyme catalyses the following chemical reaction
Galactan 5-O-arabinofuranosyltransferase is an enzyme with systematic name galactofuranan:trans,octacis-decaprenylphospho-beta-D-arabinofuranose 5-O-alpha-D-arabinofuranosyltransferase. This enzyme catalyses the following chemical reaction
Arabinofuranan 3-O-arabinosyltransferase is an enzyme with systematic name alpha-(1->5)-arabinofuranan:trans,octacis-decaprenylphospho-beta-D-arabinofuranose 3-O-alpha-D-arabinofuranosyltransferase. This enzyme catalyses the following chemical reaction:
Trans,polycis-decaprenyl diphosphate synthase is an enzyme with systematic name (2Z,6E)-farnesyl-diphosphate:isopentenyl-diphosphate farnesylcistransferase . This enzyme catalyses the following chemical reaction
UDP-N-acetylglucosamine---decaprenyl-phosphate N-acetylglucosaminephosphotransferase is an enzyme with systematic name UDP-N-acetyl-alpha-D-glucosamine:trans,octacis-decaprenyl-phosphate N-acetylglucosaminephosphotransferase. This enzyme catalyses the following chemical reaction
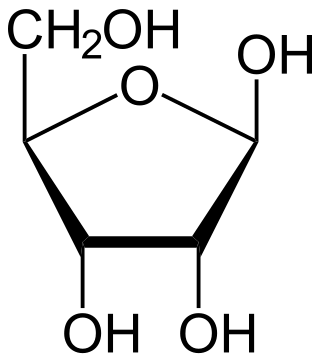
Ribose is a simple sugar and carbohydrate with molecular formula C5H10O5 and the linear-form composition H−(C=O)−(CHOH)4−H. The naturally-occurring form, d-ribose, is a component of the ribonucleotides from which RNA is built, and so this compound is necessary for coding, decoding, regulation and expression of genes. It has a structural analog, deoxyribose, which is a similarly essential component of DNA. l-ribose is an unnatural sugar that was first prepared by Emil Fischer and Oscar Piloty in 1891. It was not until 1909 that Phoebus Levene and Walter Jacobs recognised that d-ribose was a natural product, the enantiomer of Fischer and Piloty's product, and an essential component of nucleic acids. Fischer chose the name "ribose" as it is a partial rearrangement of the name of another sugar, arabinose, of which ribose is an epimer at the 2' carbon; both names also relate to gum arabic, from which arabinose was first isolated and from which they prepared l-ribose.
D-Ribose pyranase is an enzyme that catalyzes the interconversion of β-D-ribopyranose and β-D-ribofuranose. This enzyme is an isomerase that has only been found in bacteria and viruses. It has two known functions of helping transport ribose into cells and producing β-D-ribofuranose, which can later be used to make ribose 5-phosphate for the pentose phosphate pathway (PPP). D-Ribose pyranase does not have a defined crystal structure but there are two different proposed structures. The active site of D-ribose pyranase is high in histidine residues along with a few other key binding sites.








