
The citric acid cycle—also known as the Krebs cycle, Szent-Györgyi-Krebs cycle or the TCA cycle (tricarboxylic acid cycle)—is a series of biochemical reactions to release the energy stored in nutrients through the oxidation of acetyl-CoA derived from carbohydrates, fats, and proteins. The chemical energy released is available under the form of ATP. The Krebs cycle is used by organisms that respire (as opposed to organisms that ferment) to generate energy, either by anaerobic respiration or aerobic respiration. In addition, the cycle provides precursors of certain amino acids, as well as the reducing agent NADH, that are used in numerous other reactions. Its central importance to many biochemical pathways suggests that it was one of the earliest components of metabolism. Even though it is branded as a 'cycle', it is not necessary for metabolites to follow only one specific route; at least three alternative segments of the citric acid cycle have been recognized.

The Entner–Doudoroff pathway is a metabolic pathway that is most notable in Gram-negative bacteria, certain Gram-positive bacteria and archaea. Glucose is the substrate in the ED pathway and through a series of enzyme assisted chemical reactions it is catabolized into pyruvate. Entner and Doudoroff (1952) and MacGee and Doudoroff (1954) first reported the ED pathway in the bacterium Pseudomonas saccharophila. While originally thought to be just an alternative to glycolysis (EMP) and the pentose phosphate pathway (PPP), some studies now suggest that the original role of the EMP may have originally been about anabolism and repurposed over time to catabolism, meaning the ED pathway may be the older pathway. Recent studies have also shown the prevalence of the ED pathway may be more widespread than first predicted with evidence supporting the presence of the pathway in cyanobacteria, ferns, algae, mosses, and plants. Specifically, there is direct evidence that Hordeum vulgare uses the Entner–Doudoroff pathway.
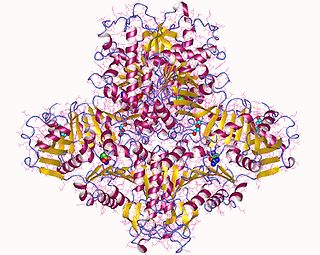
N-Acetylglutamate synthase (NAGS) is an enzyme that catalyses the production of N-acetylglutamate (NAG) from glutamate and acetyl-CoA.
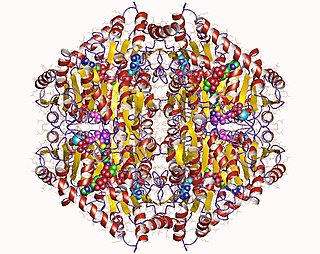
The enzyme oxalyl-CoA decarboxylase (OXC) (EC 4.1.1.8), primarily produced by the gastrointestinal bacterium Oxalobacter formigenes, catalyzes the chemical reaction

In molecular biology, the protein domain SAICAR synthase is an enzyme which catalyses a reaction to create SAICAR. In enzymology, this enzyme is also known as phosphoribosylaminoimidazolesuccinocarboxamide synthase. It is an enzyme that catalyzes the chemical reaction
The enzyme mannosyl-3-phosphoglycerate phosphatase (EC 3.1.3.70) catalyzes the reaction
In enzymology, a mannosyl-3-phosphoglycerate synthase is an enzyme that catalyzes the chemical reaction

Cobalamin biosynthesis is the process by which bacteria and archea make cobalamin, vitamin B12. Many steps are involved in converting aminolevulinic acid via uroporphyrinogen III and adenosylcobyric acid to the final forms in which it is used by enzymes in both the producing organisms and other species, including humans who acquire it through their diet.
Glucosyl-3-phosphoglycerate synthase is an enzyme with systematic name NDP-glucose:3-phospho-D-glycerate 2-alpha-D-glucosyltransferase. This enzyme catalyses the following chemical reaction
Mannosylglycerate synthase is an enzyme with systematic name GDP-mannose:D-glycerate 2-alpha-D-mannosyltransferase. This enzyme catalyses the following chemical reaction
Mannosylglucosyl-3-phosphoglycerate synthase is an enzyme with systematic name GDP-mannose:2-O-(alpha-D-glucosyl)-3-phospho-D-glycerate 2-O-alpha-D-mannosyltransferase. This enzyme catalyses the following chemical reaction
The alpha-D-phosphohexomutases are a large superfamily of enzymes, with members in all three domains of life. Enzymes from this superfamily are ubiquitous in organisms from E. coli to humans, and catalyze a phosphoryl transfer reaction on a phosphosugar substrate. Four well studied subgroups in the superfamily are:
- Phosphoglucomutase (PGM)
- Phosphoglucomutase/Phosphomannomutase (PGM/PMM)
- Phosphoglucosamine mutase (PNGM)
- Phosphoaceytlglucosamine mutase (PAGM)
1L-myo-inositol 1-phosphate cytidylyltransferase is an enzyme with systematic name CTP:1L-myo-inositol 1-phosphate cytidylyltransferase. This enzyme catalyses the following chemical reaction
Adenosylcobinamide-GDP ribazoletransferase is an enzyme with systematic name adenosylcobinamide-GDP:alpha-ribazole ribazoletransferase. This enzyme catalyses the following chemical reaction
UDP-N-acetylglucosamine—undecaprenyl-phosphate N-acetylglucosaminephosphotransferase is an enzyme with systematic name UDP-N-acetyl-alpha-D-glucosamine:ditrans,octacis-undecaprenyl phosphate N-acetyl-alpha-D-glucosaminephosphotransferase. This enzyme catalyses the following chemical reaction
CDP-L-myo-inositol myo-inositolphosphotransferase is an enzyme with systematic name CDP-1L-myo-inositol:1L-myo-inositol 1-phosphate myo-inositolphosphotransferase. This enzyme catalyses the following chemical reaction
Glucosyl-3-phosphoglycerate phosphatase (EC 3.1.3.85, GpgP protein) is an enzyme with systematic name α-D-glucosyl-3-phospho-D-glycerate phosphohydrolase. This enzyme catalyses the following chemical reaction
N2-citryl-N6-acetyl-N6-hydroxylysine synthase (EC 6.3.2.38, N(alpha)-citryl-N(epsilon)-acetyl-N(epsilon)-hydroxylysine synthase, iucA (gene)) is an enzyme with systematic name citrate:N6-acetyl-N6-hydroxy-L-lysine ligase (ADP-forming). This enzyme catalyses the following chemical reaction
Persephonella marina is a Gram-negative, rod shaped bacteria that is a member of the Aquificota phylum. Stemming from Greek, the name Persephonella is based upon the mythological goddess Persephone. Marina stems from a Latin origin, meaning "belonging to the sea". It is a thermophile with an obligate chemolithoautotrophic metabolism. Growth of P. marina can occur in pairs or individually, but is rarely seen aggregating in large groups. The organism resides on sulfidic chimneys in the deep ocean and has never been documented as a pathogen.
Rubrobacter xylanophilus is a thermophilic species of bacteria. It is slightly halotolerant, short rod- and coccus-shaped and gram-positive, with type strain PRD-1T. It is the only known true radiation resistant thermophile. It can degrade xylan and hemicellulose. The first strain of the genus Rubrobacter was isolated from gamma-irradiated hot spring water samples by Yoshinaka. This organism was found to be extremely gamma-radiation resistant, with a higher shoulder dose than the canonical radiation resistant species of the genus Deinococcus. The organism stained Gram-positive and was slightly thermophilic with an optimum growth temperature of about 60 °C.






