
A protease is an enzyme that catalyzes proteolysis, breaking down proteins into smaller polypeptides or single amino acids, and spurring the formation of new protein products. They do this by cleaving the peptide bonds within proteins by hydrolysis, a reaction where water breaks bonds. Proteases are involved in many biological functions, including digestion of ingested proteins, protein catabolism, and cell signaling.

Staphylococcus aureus is a Gram-positive spherically shaped bacterium, a member of the Bacillota, and is a usual member of the microbiota of the body, frequently found in the upper respiratory tract and on the skin. It is often positive for catalase and nitrate reduction and is a facultative anaerobe that can grow without the need for oxygen. Although S. aureus usually acts as a commensal of the human microbiota, it can also become an opportunistic pathogen, being a common cause of skin infections including abscesses, respiratory infections such as sinusitis, and food poisoning. Pathogenic strains often promote infections by producing virulence factors such as potent protein toxins, and the expression of a cell-surface protein that binds and inactivates antibodies. S. aureus is one of the leading pathogens for deaths associated with antimicrobial resistance and the emergence of antibiotic-resistant strains, such as methicillin-resistant S. aureus (MRSA), is a worldwide problem in clinical medicine. Despite much research and development, no vaccine for S. aureus has been approved.

Serine proteases are enzymes that cleave peptide bonds in proteins. Serine serves as the nucleophilic amino acid at the (enzyme's) active site. They are found ubiquitously in both eukaryotes and prokaryotes. Serine proteases fall into two broad categories based on their structure: chymotrypsin-like (trypsin-like) or subtilisin-like.

Cysteine proteases, also known as thiol proteases, are hydrolase enzymes that degrade proteins. These proteases share a common catalytic mechanism that involves a nucleophilic cysteine thiol in a catalytic triad or dyad.
Virulence factors are cellular structures, molecules and regulatory systems that enable microbial pathogens to achieve the following:

Panton–Valentine leukocidin (PVL) is a cytotoxin—one of the β-pore-forming toxins. The presence of PVL is associated with increased virulence of certain strains (isolates) of Staphylococcus aureus. It is present in the majority of community-associated methicillin-resistant Staphylococcus aureus (CA-MRSA) isolates studied and is the cause of necrotic lesions involving the skin or mucosa, including necrotic hemorrhagic pneumonia. PVL creates pores in the membranes of infected cells. PVL is produced from the genetic material of a bacteriophage that infects Staphylococcus aureus, making it more virulent.

Hemolysins or haemolysins are lipids and proteins that cause lysis of red blood cells by disrupting the cell membrane. Although the lytic activity of some microbe-derived hemolysins on red blood cells may be of great importance for nutrient acquisition, many hemolysins produced by pathogens do not cause significant destruction of red blood cells during infection. However, hemolysins are often capable of lysing red blood cells in vitro.
In molecular biology SprD is a non-coding RNA expressed on pathogenicity islands in Staphylococcus aureus. It was identified in silico along with a number of other sRNAs (SprA-G) through microarray analysis which were confirmed using a Northern blot. SprD has been found to significantly contribute to causing disease in an animal model.

Streptococcal pyrogenic exotoxins also known as erythrogenic toxins, are exotoxins secreted by strains of the bacterial species Streptococcus pyogenes. SpeA and speC are superantigens, which induce inflammation by nonspecifically activating T cells and stimulating the production of inflammatory cytokines. SpeB, the most abundant streptococcal extracellular protein, is a cysteine protease. Pyrogenic exotoxins are implicated as the causative agent of scarlet fever and streptococcal toxic shock syndrome. There is no consensus on the exact number of pyrogenic exotoxins. Serotypes A-C are the most extensively studied and recognized by all sources, but others note up to thirteen distinct types, categorizing speF through speM as additional superantigens. Erythrogenic toxins are known to damage the plasma membranes of blood capillaries under the skin and produce a red skin rash. Past studies have shown that multiple variants of erythrogenic toxins may be produced, depending on the strain of S. pyogenes in question. Some strains may not produce a detectable toxin at all. Bacteriophage T12 infection of S. pyogenes enables the production of speA, and increases virulence.

OmpT is an aspartyl protease found on the outer membrane of Escherichia coli. OmpT is a subtype of the family of omptin proteases, which are found on some gram-negative species of bacteria.
Staphopain is an enzyme. This enzyme catalyses the following chemical reaction

Aureolysin is an extracellular metalloprotease expressed by Staphylococcus aureus. This protease is a major contributor to the bacterium's virulence, or ability to cause disease, by cleaving host factors of the innate immune system as well as regulating S. aureus secreted toxins and cell wall proteins. To catalyze its enzymatic activities, aureolysin requires zinc and calcium which it obtains from the extracellular environment within the host.
Staphylococcus pseudintermedius is a gram positive coccus bacteria of the genus Staphylococcus found worldwide. It is primarily a pathogen for domestic animals, but has been known to affect humans as well. S. pseudintermedius is an opportunistic pathogen that secretes immune modulating virulence factors, has many adhesion factors, and the potential to create biofilms, all of which help to determine the pathogenicity of the bacterium. Diagnoses of Staphylococcus pseudintermedius have traditionally been made using cytology, plating, and biochemical tests. More recently, molecular technologies like MALDI-TOF, DNA hybridization and PCR have become preferred over biochemical tests for their more rapid and accurate identifications. This includes the identification and diagnosis of antibiotic resistant strains.

The PA clan is the largest group of proteases with common ancestry as identified by structural homology. Members have a chymotrypsin-like fold and similar proteolysis mechanisms but can have identity of <10%. The clan contains both cysteine and serine proteases. PA clan proteases can be found in plants, animals, fungi, eubacteria, archaea and viruses.
Glutamyl endopeptidase I is a family of extracellular bacterial serine proteases. The proteases within this family have been identified in species of Staphylococcus, Bacillus, and Streptomyces, among others. The two former are more closely related, while the Streptomyces-type is treated as a separate family, glutamyl endopeptidase II.
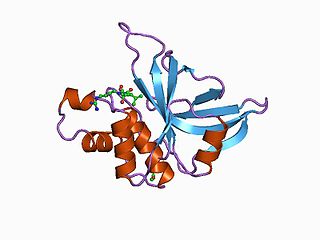
Staphopain A is a secreted cysteine protease produced by Staphylococcus aureus. It was first identified in the S. aureus V8 strain as a papain-like cysteine protease. The protease distinguishes itself from the other major proteases of S. aureus in its very broad specificity and its ability to degrade elastin.

In the contact activation system or CAS, three proteins in the blood, factor XII (FXII), prekallikrein (PK) and high molecular weight kininogen (HK), bind to a surface and cause blood coagulation and inflammation. FXII and PK are proteases and HK is a non-enzymatic co-factor. The CAS can activate the kinin–kallikrein system and blood coagulation through its ability to activate multiple downstream proteins. The CAS is initiated when FXII binds to a surface and reciprocal activation of FXII and PK occurs, forming FXIIa and PKa. FXIIa can initiate the coagulation cascade by cleaving and activating factor XI (FXI), which leads to formation of a blood clot. Additionally, the CAS can activate the kinin–kallikrein system when PKa cleaves HK to form cHK, releasing a peptide known as bradykinin (BK). BK and its derivatives bind to bradykinin receptors B1 and B2 to mediate inflammation.
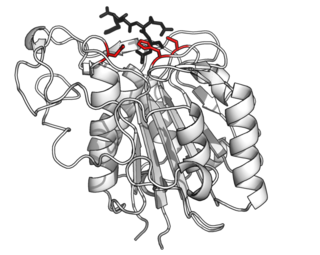
Asparagine endopeptidase is a proteolytic enzyme from C13 peptidase family which hydrolyses a peptide bond using the thiol group of a cysteine residue as a nucleophile. It is also known as asparaginyl endopeptidase, citvac, proteinase B, hemoglobinase, PRSC1 gene product or LGMN, vicilin peptidohydrolase and bean endopeptidase. In humans it is encoded by the LGMN gene.
Accessory gene regulator (agr) is a complex 5 gene locus that is a global regulator of virulence in Staphylococcus aureus. It encodes a two-component transcriptional quorum-sensing (QS) system activated by an autoinducing, thiolactone-containing cyclic peptide (AIP).

Papain-like proteases are a large protein family of cysteine protease enzymes that share structural and enzymatic properties with the group's namesake member, papain. They are found in all domains of life. In animals, the group is often known as cysteine cathepsins or, in older literature, lysosomal peptidases. In the MEROPS protease enzyme classification system, papain-like proteases form Clan CA. Papain-like proteases share a common catalytic dyad active site featuring a cysteine amino acid residue that acts as a nucleophile.















