
Ribonuclease is a type of nuclease that catalyzes the degradation of RNA into smaller components. Ribonucleases can be divided into endoribonucleases and exoribonucleases, and comprise several sub-classes within the EC 2.7 and 3.1 classes of enzymes.

Ribonuclease H is a family of non-sequence-specific endonuclease enzymes that catalyze the cleavage of RNA in an RNA/DNA substrate via a hydrolytic mechanism. Members of the RNase H family can be found in nearly all organisms, from bacteria to archaea to eukaryotes.

Dicer, also known as endoribonuclease Dicer or helicase with RNase motif, is an enzyme that in humans is encoded by the DICER1 gene. Being part of the RNase III family, Dicer cleaves double-stranded RNA (dsRNA) and pre-microRNA (pre-miRNA) into short double-stranded RNA fragments called small interfering RNA and microRNA, respectively. These fragments are approximately 20–25 base pairs long with a two-base overhang on the 3′-end. Dicer facilitates the activation of the RNA-induced silencing complex (RISC), which is essential for RNA interference. RISC has a catalytic component Argonaute, which is an endonuclease capable of degrading messenger RNA (mRNA).
In molecular biology, endonucleases are enzymes that cleave the phosphodiester bond within a polynucleotide chain. Some, such as deoxyribonuclease I, cut DNA relatively nonspecifically, while many, typically called restriction endonucleases or restriction enzymes, cleave only at very specific nucleotide sequences. Endonucleases differ from exonucleases, which cleave the ends of recognition sequences instead of the middle (endo) portion. Some enzymes known as "exo-endonucleases", however, are not limited to either nuclease function, displaying qualities that are both endo- and exo-like. Evidence suggests that endonuclease activity experiences a lag compared to exonuclease activity.
The RNA-induced silencing complex, or RISC, is a multiprotein complex, specifically a ribonucleoprotein, which functions in gene silencing via a variety of pathways at the transcriptional and translational levels. Using single-stranded RNA (ssRNA) fragments, such as microRNA (miRNA), or double-stranded small interfering RNA (siRNA), the complex functions as a key tool in gene regulation. The single strand of RNA acts as a template for RISC to recognize complementary messenger RNA (mRNA) transcript. Once found, one of the proteins in RISC, Argonaute, activates and cleaves the mRNA. This process is called RNA interference (RNAi) and it is found in many eukaryotes; it is a key process in defense against viral infections, as it is triggered by the presence of double-stranded RNA (dsRNA).

Ribonuclease L or RNase L, known sometimes as ribonuclease 4 or 2'-5' oligoadenylate synthetase-dependent ribonuclease, is an interferon (IFN)-induced ribonuclease which, upon activation, destroys all RNA within the cell. RNase L is an enzyme that in humans is encoded by the RNASEL gene.

Angiogenin (ANG) also known as ribonuclease 5 is a small 123 amino acid protein that in humans is encoded by the ANG gene. Angiogenin is a potent stimulator of new blood vessels through the process of angiogenesis. Ang hydrolyzes cellular RNA, resulting in modulated levels of protein synthesis and interacts with DNA causing a promoter-like increase in the expression of rRNA. Ang is associated with cancer and neurological disease through angiogenesis and through activating gene expression that suppresses apoptosis.
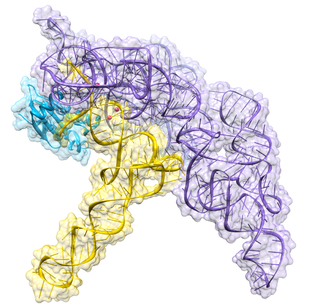
Ribonuclease P is a type of ribonuclease which cleaves RNA. RNase P is unique from other RNases in that it is a ribozyme – a ribonucleic acid that acts as a catalyst in the same way that a protein-based enzyme would. Its function is to cleave off an extra, or precursor, sequence of RNA on tRNA molecules. Further, RNase P is one of two known multiple turnover ribozymes in nature, the discovery of which earned Sidney Altman and Thomas Cech the Nobel Prize in Chemistry in 1989: in the 1970s, Altman discovered the existence of precursor tRNA with flanking sequences and was the first to characterize RNase P and its activity in processing of the 5' leader sequence of precursor tRNA. Recent findings also reveal that RNase P has a new function. It has been shown that human nuclear RNase P is required for the normal and efficient transcription of various small noncoding RNAs, such as tRNA, 5S rRNA, SRP RNA and U6 snRNA genes, which are transcribed by RNA polymerase III, one of three major nuclear RNA polymerases in human cells.

The Argonaute protein family, first discovered for its evolutionarily conserved stem cell function, plays a central role in RNA silencing processes as essential components of the RNA-induced silencing complex (RISC). RISC is responsible for the gene silencing phenomenon known as RNA interference (RNAi). Argonaute proteins bind different classes of small non-coding RNAs, including microRNAs (miRNAs), small interfering RNAs (siRNAs) and Piwi-interacting RNAs (piRNAs). Small RNAs guide Argonaute proteins to their specific targets through sequence complementarity, which then leads to mRNA cleavage, translation inhibition, and/or the initiation of mRNA decay.

Drosha is a Class 2 ribonuclease III enzyme that in humans is encoded by the DROSHA gene. It is the primary nuclease that executes the initiation step of miRNA processing in the nucleus. It works closely with DGCR8 and in correlation with Dicer. It has been found significant in clinical knowledge for cancer prognosis and HIV-1 replication.
esiRNA or Endoribonuclease-prepared siRNAs are a mixture of siRNA oligos resulting from cleavage of long double-stranded RNA (dsRNA) with an endoribonuclease such as Escherichia coli RNase III or Dicer.
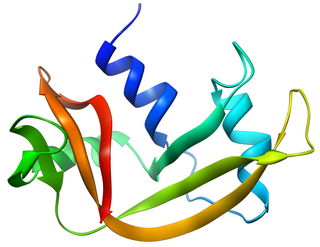
Pancreatic ribonuclease family is a superfamily of pyrimidine-specific endonucleases found in high quantity in the pancreas of certain mammals and of some reptiles.
The degradosome is a multiprotein complex present in most bacteria that is involved in the processing of ribosomal RNA and the degradation of messenger RNA and is regulated by Non-coding RNA. It contains the proteins RNA helicase B, RNase E and Polynucleotide phosphorylase.

Ribonuclease H1 also known as RNase H1 is an enzyme that in humans is encoded by the RNASEH1 gene. The RNase H1 is a non-specific endonuclease and catalyzes the cleavage of RNA via a hydrolytic mechanism.

Ribonuclease H2 subunit A, also known as RNase H2 subunit A, is an enzyme that in humans is encoded by the RNASEH2A gene.
Ribonuclease E is a bacterial ribonuclease that participates in the processing of ribosomal RNA and the chemical degradation of bulk cellular RNA.
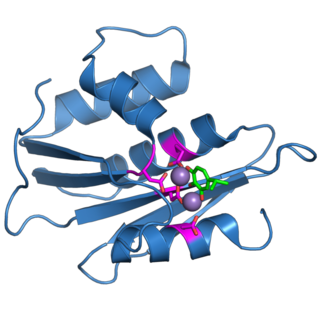
The retroviral ribonuclease H is a catalytic domain of the retroviral reverse transcriptase (RT) enzyme. The RT enzyme is used to generate complementary DNA (cDNA) from the retroviral RNA genome. This process is called reverse transcription. To complete this complex process, the retroviral RT enzymes need to adopt a multifunctional nature. They therefore possess 3 of the following biochemical activities: RNA-dependent DNA polymerase, ribonuclease H, and DNA-dependent DNA polymerase activities. Like all RNase H enzymes, the retroviral RNase H domain cleaves DNA/RNA duplexes and will not degrade DNA or unhybridized RNA.

The microprocessor complex is a protein complex involved in the early stages of processing microRNA (miRNA) and RNA interference (RNAi) in animal cells. The complex is minimally composed of the ribonuclease enzyme Drosha and the dimeric RNA-binding protein DGCR8, and cleaves primary miRNA substrates to pre-miRNA in the cell nucleus. Microprocessor is also the smaller of the two multi-protein complexes that contain human Drosha.

DCL1 is a gene in plants that codes for the DCL1 protein, a ribonuclease III enzyme involved in processing double-stranded RNA (dsRNA) and microRNA (miRNA). Although DCL1, also called Endoribonuclease Dicer homolog 1, is named for its homology with the metazoan protein Dicer, its role in miRNA biogenesis is somewhat different, due to substantial differences in miRNA maturation processes between plants and animals, as well due to additional downstream plant-specific pathways, where DCL1 paralogs like DCL4 participate, such Trans-acting siRNA biogenesis.
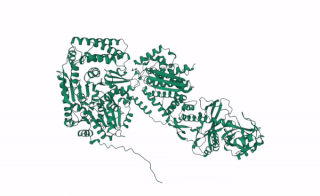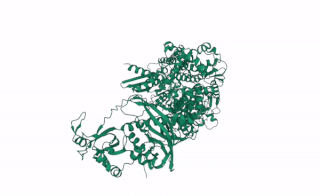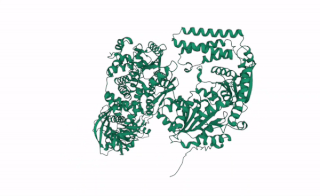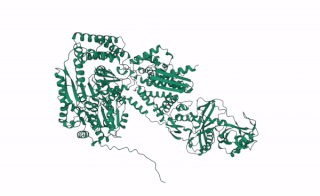
DCL2 is a gene in plants that codes for the DCL2 protein, a ribonuclease III enzyme involved in processing exogenous double-stranded RNA (dsRNA) into 22 nucleotide small interference RNAs (siRNAs).

















