Related Research Articles

Endocytosis is a cellular process in which substances are brought into the cell. The material to be internalized is surrounded by an area of cell membrane, which then buds off inside the cell to form a vesicle containing the ingested materials. Endocytosis includes pinocytosis and phagocytosis. It is a form of active transport.
GTPases are a large family of hydrolase enzymes that bind to the nucleotide guanosine triphosphate (GTP) and hydrolyze it to guanosine diphosphate (GDP). The GTP binding and hydrolysis takes place in the highly conserved P-loop "G domain", a protein domain common to many GTPases.
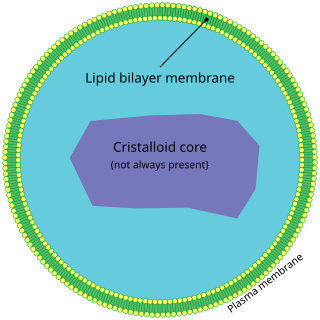
A peroxisome (IPA:[pɛɜˈɹɒksɪˌsoʊm]) is a membrane-bound organelle, a type of microbody, found in the cytoplasm of virtually all eukaryotic cells. Peroxisomes are oxidative organelles. Frequently, molecular oxygen serves as a co-substrate, from which hydrogen peroxide (H2O2) is then formed. Peroxisomes owe their name to hydrogen peroxide generating and scavenging activities. They perform key roles in lipid metabolism and the reduction of reactive oxygen species.
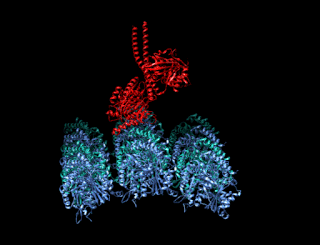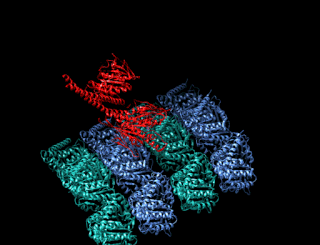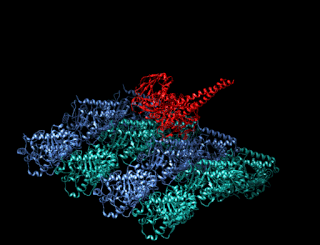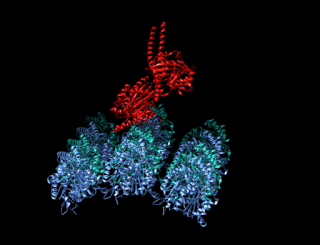
A kinesin is a protein belonging to a class of motor proteins found in eukaryotic cells. Kinesins move along microtubule (MT) filaments and are powered by the hydrolysis of adenosine triphosphate (ATP). The active movement of kinesins supports several cellular functions including mitosis, meiosis and transport of cellular cargo, such as in axonal transport, and intraflagellar transport. Most kinesins walk towards the plus end of a microtubule, which, in most cells, entails transporting cargo such as protein and membrane components from the center of the cell towards the periphery. This form of transport is known as anterograde transport. In contrast, dyneins are motor proteins that move toward the minus end of a microtubule in retrograde transport.
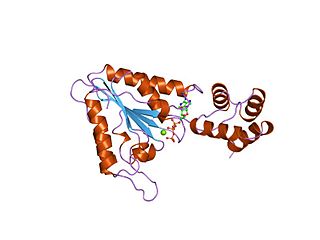
AAAproteins are a large group of protein family sharing a common conserved module of approximately 230 amino acid residues. This is a large, functionally diverse protein family belonging to the AAA+ protein superfamily of ring-shaped P-loop NTPases, which exert their activity through the energy-dependent remodeling or translocation of macromolecules.
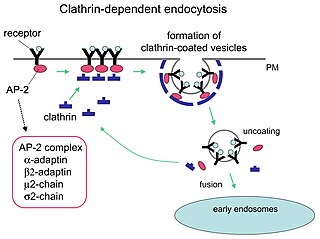
Receptor-mediated endocytosis (RME), also called clathrin-mediated endocytosis, is a process by which cells absorb metabolites, hormones, proteins – and in some cases viruses – by the inward budding of the plasma membrane (invagination). This process forms vesicles containing the absorbed substances and is strictly mediated by receptors on the surface of the cell. Only the receptor-specific substances can enter the cell through this process.

A fusion gene is a hybrid gene formed from two previously independent genes. It can occur as a result of translocation, interstitial deletion, or chromosomal inversion. Fusion genes have been found to be prevalent in all main types of human neoplasia. The identification of these fusion genes play a prominent role in being a diagnostic and prognostic marker.
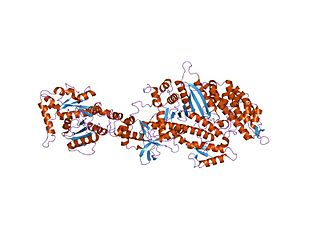
Dynamin is a GTPase responsible for endocytosis in the eukaryotic cell. Dynamin is part of the "dynamin superfamily", which includes classical dynamins, dynamin-like proteins, Mx proteins, OPA1, mitofusins, and GBPs. Members of the dynamin family are principally involved in the scission of newly formed vesicles from the membrane of one cellular compartment and their targeting to, and fusion with, another compartment, both at the cell surface as well as at the Golgi apparatus. Dynamin family members also play a role in many processes including division of organelles, cytokinesis and microbial pathogen resistance.

The immunoglobulin superfamily (IgSF) is a large protein superfamily of cell surface and soluble proteins that are involved in the recognition, binding, or adhesion processes of cells. Molecules are categorized as members of this superfamily based on shared structural features with immunoglobulins ; they all possess a domain known as an immunoglobulin domain or fold. Members of the IgSF include cell surface antigen receptors, co-receptors and co-stimulatory molecules of the immune system, molecules involved in antigen presentation to lymphocytes, cell adhesion molecules, certain cytokine receptors and intracellular muscle proteins. They are commonly associated with roles in the immune system. Otherwise, the sperm-specific protein IZUMO1, a member of the immunoglobulin superfamily, has also been identified as the only sperm membrane protein essential for sperm-egg fusion.
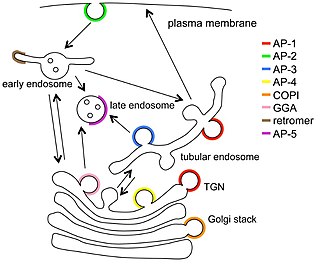
Vesicular transport adaptor proteins are proteins involved in forming complexes that function in the trafficking of molecules from one subcellular location to another. These complexes concentrate the correct cargo molecules in vesicles that bud or extrude off of one organelle and travel to another location, where the cargo is delivered. While some of the details of how these adaptor proteins achieve their trafficking specificity has been worked out, there is still much to be learned.

Mitofusin-2 is a protein that in humans is encoded by the MFN2 gene. Mitofusins are GTPases embedded in the outer membrane of the mitochondria. In mammals MFN1 and MFN2 are essential for mitochondrial fusion. In addition to the mitofusins, OPA1 regulates inner mitochondrial membrane fusion, and DRP1 is responsible for mitochondrial fission.

Dynamin-like 120 kDa protein, mitochondrial is a protein that in humans is encoded by the OPA1 gene. This protein regulates mitochondrial fusion and cristae structure in the inner mitochondrial membrane (IMM) and contributes to ATP synthesis and apoptosis, and small, round mitochondria. Mutations in this gene have been implicated in dominant optic atrophy (DOA), leading to loss in vision, hearing, muscle contraction, and related dysfunctions.

Dynamin-1-like protein is a GTPase that regulates mitochondrial fission. In humans, dynamin-1-like protein, which is typically referred to as dynamin-related protein 1 (Drp1), is encoded by the DNM1L gene and is part of the dynamin superfamily (DSP) family of proteins.

Mitochondrial fission 1 protein (FIS1) is a protein that in humans is encoded by the FIS1 gene on chromosome 7. This protein is a component of a mitochondrial complex, the ARCosome, that promotes mitochondrial fission. Its role in mitochondrial fission thus implicates it in the regulation of mitochondrial morphology, the cell cycle, and apoptosis. By extension, the protein is involved in associated diseases, including neurodegenerative diseases and cancers.
Mitochondrial biogenesis is the process by which cells increase mitochondrial numbers. It was first described by John Holloszy in the 1960s, when it was discovered that physical endurance training induced higher mitochondrial content levels, leading to greater glucose uptake by muscles. Mitochondrial biogenesis is activated by numerous different signals during times of cellular stress or in response to environmental stimuli, such as aerobic exercise.
The EHD protein family is a relatively small group of proteins which have been shown to play a role in several physiological functions, the most notable being the regulation of endocytotic vesicles. This family is recognized by its highly conserved EH domain, a structural motif that has been shown to facilitate specificity and interaction between protein and ligand. The four mammalian EHD proteins that have been classified are: EHD1, EHD2, EHD3, and EHD4.

A circular permutation is a relationship between proteins whereby the proteins have a changed order of amino acids in their peptide sequence. The result is a protein structure with different connectivity, but overall similar three-dimensional (3D) shape. In 1979, the first pair of circularly permuted proteins – concanavalin A and lectin – were discovered; over 2000 such proteins are now known.
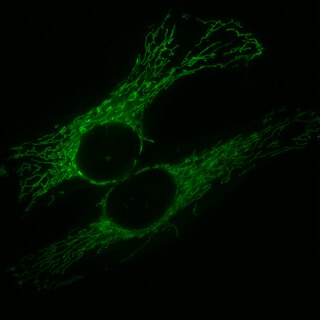
Mitochondrial fission is the process where mitochondria divide or segregate into two separate mitochondrial organelles. Mitochondrial fission is counteracted by the process of mitochondrial fusion, whereby two separate mitochondria can fuse together to form a large one. Mitochondrial fusion in turn can result in elongated mitochondrial networks. Both mitochondrial fission and fusion are balanced in the cell, and mutations interfering with either processes are associated with a variety of diseases. Mitochondria can divide by prokaryotic binary fission and since they require mitochondrial DNA for their function, fission is coordinated with DNA replication. Some of the proteins that are involved in mitochondrial fission have been identified and some of them are associated with mitochondrial diseases. Mitochondrial fission has significant implications in stress response and apoptosis.

Mitochondria are dynamic organelles with the ability to fuse and divide (fission), forming constantly changing tubular networks in most eukaryotic cells. These mitochondrial dynamics, first observed over a hundred years ago are important for the health of the cell, and defects in dynamics lead to genetic disorders. Through fusion, mitochondria can overcome the dangerous consequences of genetic malfunction. The process of mitochondrial fusion involves a variety of proteins that assist the cell throughout the series of events that form this process.

Gia Voeltz is an American cell biologist. She is a professor of Molecular, Cellular and Developmental Biology at the University of Colorado Boulder and a Howard Hughes Medical Institute Investigator. She is known for her research identifying the factors and unraveling the mechanisms that determine the structure and dynamics of the largest organelle in the cell: the endoplasmic reticulum. Her lab has produced paradigm shifting studies on organelle membrane contact sites that have revealed that most cytoplasmic organelles are not isolated entities but are instead physically tethered to an interconnected ER membrane network.
References
- ↑ Henley JR, Cao H, McNiven MA (December 1999). "Participation of dynamin in the biogenesis of cytoplasmic vesicles". FASEB Journal. 13 Suppl 2 (9002): S243-7. doi: 10.1096/fasebj.13.9002.S243 . PMID 10619136. S2CID 24401725.
- 1 2 Jimah, JR; Hinshaw, JE (March 2019). "Structural Insights into the Mechanism of Dynamin Superfamily Proteins". Trends in Cell Biology. 29 (3): 257–273. doi:10.1016/j.tcb.2018.11.003. PMC 9623552 . PMID 30527453. S2CID 54478789.