The enzyme Uridine monophosphate synthase catalyses the formation of uridine monophosphate (UMP), an energy-carrying molecule in many important biosynthetic pathways. In humans, the gene that codes for this enzyme is located on the long arm of chromosome 3 (3q13).
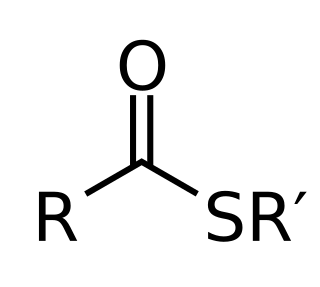
In organic chemistry, thioesters are organosulfur compounds with the molecular structure R−C(=O)−S−R’. They are analogous to carboxylate esters with the sulfur in the thioester replacing oxygen in the carboxylate ester, as implied by the thio- prefix. They are the product of esterification of a carboxylic acid with a thiol. In biochemistry, the best-known thioesters are derivatives of coenzyme A, e.g., acetyl-CoA. The R and R' represent organyl groups, or H in the case of R.

Aromatic L-amino acid decarboxylase, also known as DOPA decarboxylase (DDC), tryptophan decarboxylase, and 5-hydroxytryptophan decarboxylase, is a lyase enzyme, located in region 7p12.2-p12.1.

Pyridoxal phosphate (PLP, pyridoxal 5'-phosphate, P5P), the active form of vitamin B6, is a coenzyme in a variety of enzymatic reactions. The International Union of Biochemistry and Molecular Biology has catalogued more than 140 PLP-dependent activities, corresponding to ~4% of all classified activities. The versatility of PLP arises from its ability to covalently bind the substrate, and then to act as an electrophilic catalyst, thereby stabilizing different types of carbanionic reaction intermediates.

A catalytic triad is a set of three coordinated amino acids that can be found in the active site of some enzymes. Catalytic triads are most commonly found in hydrolase and transferase enzymes. An acid-base-nucleophile triad is a common motif for generating a nucleophilic residue for covalent catalysis. The residues form a charge-relay network to polarise and activate the nucleophile, which attacks the substrate, forming a covalent intermediate which is then hydrolysed to release the product and regenerate free enzyme. The nucleophile is most commonly a serine or cysteine amino acid, but occasionally threonine or even selenocysteine. The 3D structure of the enzyme brings together the triad residues in a precise orientation, even though they may be far apart in the sequence.

Methionine synthase (MS, MeSe, MTR) is responsible for the regeneration of methionine from homocysteine. In humans it is encoded by the MTR gene (5-methyltetrahydrofolate-homocysteine methyltransferase). Methionine synthase forms part of the S-adenosylmethionine (SAMe) biosynthesis and regeneration cycle, and is the enzyme responsible for linking the cycle to one-carbon metabolism via the folate cycle. There are two primary forms of this enzyme, the Vitamin B12 (cobalamin)-dependent (MetH) and independent (MetE) forms, although minimal core methionine synthases that do not fit cleanly into either category have also been described in some anaerobic bacteria. The two dominant forms of the enzymes appear to be evolutionary independent and rely on considerably different chemical mechanisms. Mammals and other higher eukaryotes express only the cobalamin-dependent form. In contrast, the distribution of the two forms in Archaeplastida (plants and algae) is more complex. Plants exclusively possess the cobalamin-independent form, while algae have either one of the two, depending on species. Many different microorganisms express both the cobalamin-dependent and cobalamin-independent forms.

Spermidine synthase is an enzyme that catalyzes the transfer of the propylamine group from S-adenosylmethioninamine to putrescine in the biosynthesis of spermidine. The systematic name is S-adenosyl 3-(methylthio)propylamine:putrescine 3-aminopropyltransferase and it belongs to the group of aminopropyl transferases. It does not need any cofactors. Most spermidine synthases exist in solution as dimers.
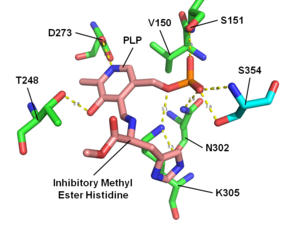
The enzyme histidine decarboxylase is transcribed on chromosome 15, region q21.1-21.2, and catalyzes the decarboxylation of histidine to form histamine. In mammals, histamine is an important biogenic amine with regulatory roles in neurotransmission, gastric acid secretion and immune response. Histidine decarboxylase is the sole member of the histamine synthesis pathway, producing histamine in a one-step reaction. Histamine cannot be generated by any other known enzyme. HDC is therefore the primary source of histamine in most mammals and eukaryotes. The enzyme employs a pyridoxal 5'-phosphate (PLP) cofactor, in similarity to many amino acid decarboxylases. Eukaryotes, as well as gram-negative bacteria share a common HDC, while gram-positive bacteria employ an evolutionarily unrelated pyruvoyl-dependent HDC. In humans, histidine decarboxylase is encoded by the HDC gene.

Oxaloacetate decarboxylase is a carboxy-lyase involved in the conversion of oxaloacetate into pyruvate.
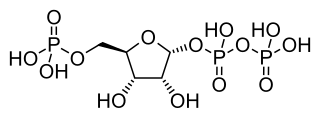
Phosphoribosyl pyrophosphate (PRPP) is a pentose phosphate. It is a biochemical intermediate in the formation of purine nucleotides via inosine-5-monophosphate, as well as in pyrimidine nucleotide formation. Hence it is a building block for DNA and RNA. The vitamins thiamine and cobalamin, and the amino acid tryptophan also contain fragments derived from PRPP. It is formed from ribose 5-phosphate (R5P) by the enzyme ribose-phosphate diphosphokinase:

Orotidine 5'-phosphate decarboxylase or orotidylate decarboxylase is an enzyme involved in pyrimidine biosynthesis. It catalyzes the decarboxylation of orotidine monophosphate (OMP) to form uridine monophosphate (UMP). The function of this enzyme is essential to the de novo biosynthesis of the pyrimidine nucleotides uridine triphosphate, cytidine triphosphate, and thymidine triphosphate. OMP decarboxylase has been a frequent target for scientific investigation because of its demonstrated extreme catalytic efficiency and its usefulness as a selection marker for yeast strain engineering.

Histidine ammonia-lyase is an enzyme that in humans is encoded by the HAL gene. It converts histidine into ammonia and urocanic acid. Its systematic name is L-histidine ammonia-lyase (urocanate-forming).
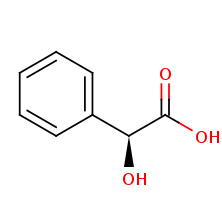
In enzymology, (S)-mandelate dehydrogenase (MDH), is an enzyme that catalyzes the chemical reaction.
The enzyme 3-dehydro-L-gulonate-6-phosphate decarboxylase (EC 4.1.1.85) catalyzes the chemical reaction

The enzyme Acid-Induced Arginine Decarboxylase (AdiA), also commonly referred to as arginine decarboxylase, catalyzes the conversion of L-arginine into agmatine and carbon dioxide. The process consumes a proton in the decarboxylation and employs a pyridoxal-5'-phosphate (PLP) cofactor, similar to other enzymes involved in amino acid metabolism, such as ornithine decarboxylase and glutamine decarboxylase. It is found in bacteria and virus, though most research has so far focused on forms of the enzyme in bacteria. During the AdiA catalyzed decarboxylation of arginine, the necessary proton is consumed from the cell cytoplasm which helps to prevent the over-accumulation of protons inside the cell and serves to increase the intracellular pH. Arginine decarboxylase is part of an enzymatic system in Escherichia coli, Salmonella Typhimurium, and methane-producing bacteria Methanococcus jannaschii that makes these organisms acid resistant and allows them to survive under highly acidic medium.

Acetylcholinesterase (HGNC symbol ACHE; EC 3.1.1.7; systematic name acetylcholine acetylhydrolase), also known as AChE, AChase or acetylhydrolase, is the primary cholinesterase in the body. It is an enzyme that catalyzes the breakdown of acetylcholine and some other choline esters that function as neurotransmitters:
Biotin-independent malonate decarboxylase (EC 4.1.1.88, malonate decarboxylase (without biotin), malonate decarboxylase, MDC) is an enzyme with systematic name malonate carboxy-lyase (biotin-independent). This enzyme catalyses the following chemical reaction

Chlorophyllide a and Chlorophyllide b are the biosynthetic precursors of chlorophyll a and chlorophyll b respectively. Their propionic acid groups are converted to phytyl esters by the enzyme chlorophyll synthase in the final step of the pathway. Thus the main interest in these chemical compounds has been in the study of chlorophyll biosynthesis in plants, algae and cyanobacteria. Chlorophyllide a is also an intermediate in the biosynthesis of bacteriochlorophylls.
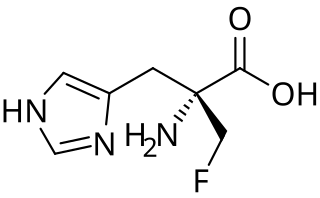
α-Fluoromethylhistidine (α-FMH) is an irreversible specific inhibitor of histidine decarboxylase (HDC). It functions by forming a covalent linkage with a catalytic serine residue on the active site of HDC. Due to its efficacy in reducing histamine levels in tissue mast cells, it has many applications in the study of histaminergic systems.
Beverly Marie Guirard was a microbiologist who worked on the biochemistry of microbial growth, especially with respect to vitamin B6. She is also known for her work defining the components of coenzyme A which was a part of the research that led to a Nobel Prize for Fritz Albert Lipmann.