
Nucleotides are organic molecules composed of a nitrogenous base, a pentose sugar and a phosphate. They serve as monomeric units of the nucleic acid polymers – deoxyribonucleic acid (DNA) and ribonucleic acid (RNA), both of which are essential biomolecules within all life-forms on Earth. Nucleotides are obtained in the diet and are also synthesized from common nutrients by the liver.
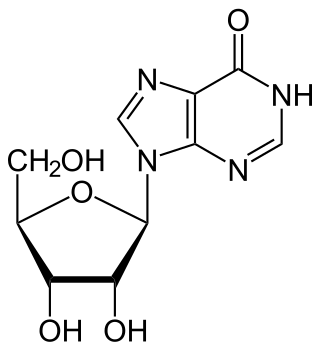
Inosine is a nucleoside that is formed when hypoxanthine is attached to a ribose ring (also known as a ribofuranose) via a β-N9-glycosidic bond. It was discovered in 1965 in analysis of RNA transferase. Inosine is commonly found in tRNAs and is essential for proper translation of the genetic code in wobble base pairs.

In biochemistry, a ribonucleotide is a nucleotide containing ribose as its pentose component. It is considered a molecular precursor of nucleic acids. Nucleotides are the basic building blocks of DNA and RNA. Ribonucleotides themselves are basic monomeric building blocks for RNA. Deoxyribonucleotides, formed by reducing ribonucleotides with the enzyme ribonucleotide reductase (RNR), are essential building blocks for DNA. There are several differences between DNA deoxyribonucleotides and RNA ribonucleotides. Successive nucleotides are linked together via phosphodiester bonds.

Mycophenolic acid is an immunosuppressant medication used to prevent rejection following organ transplantation and to treat autoimmune conditions such as Crohn's disease and lupus. Specifically it is used following kidney, heart, and liver transplantation. It can be given by mouth or by injection into a vein. It comes as mycophenolate sodium and mycophenolate mofetil.
A nucleoside triphosphate is a nucleoside containing a nitrogenous base bound to a 5-carbon sugar, with three phosphate groups bound to the sugar. They are the molecular precursors of both DNA and RNA, which are chains of nucleotides made through the processes of DNA replication and transcription. Nucleoside triphosphates also serve as a source of energy for cellular reactions and are involved in signalling pathways.

Hypoxanthine-guanine phosphoribosyltransferase (HGPRT) is an enzyme encoded in humans by the HPRT1 gene.

Inosinic acid or inosine monophosphate (IMP) is a nucleotide. Widely used as a flavor enhancer, it is typically obtained from chicken byproducts or other meat industry waste. Inosinic acid is important in metabolism. It is the ribonucleotide of hypoxanthine and the first nucleotide formed during the synthesis of purine nucleotides. It can also be formed by the deamination of adenosine monophosphate by AMP deaminase. It can be hydrolysed to inosine.
Purine nucleoside phosphorylase, PNP, PNPase or inosine phosphorylase is an enzyme that in humans is encoded by the NP gene. It catalyzes the chemical reaction

Nucleic acid metabolism is a collective term that refers to the variety of chemical reactions by which nucleic acids are either synthesized or degraded. Nucleic acids are polymers made up of a variety of monomers called nucleotides. Nucleotide synthesis is an anabolic mechanism generally involving the chemical reaction of phosphate, pentose sugar, and a nitrogenous base. Degradation of nucleic acids is a catabolic reaction and the resulting parts of the nucleotides or nucleobases can be salvaged to recreate new nucleotides. Both synthesis and degradation reactions require multiple enzymes to facilitate the event. Defects or deficiencies in these enzymes can lead to a variety of diseases.
Purine metabolism refers to the metabolic pathways to synthesize and break down purines that are present in many organisms.

Xanthosine monophosphate (xanthylate) is an intermediate in purine metabolism. It is a ribonucleoside monophosphate. It is formed from IMP via the action of IMP dehydrogenase, and it forms GMP via the action of GMP synthase. Also, XMP can be released from XTP by enzyme deoxyribonucleoside triphosphate pyrophosphohydrolase containing (d)XTPase activity.

Guanosine monophosphate synthetase, also known as GMPS is an enzyme that converts xanthosine monophosphate to guanosine monophosphate.
GMP reductase EC 1.7.1.7 is an enzyme that catalyzes the irreversible and NADPH-dependent reductive deamination of GMP into IMP.
In enzymology, an adenosine-phosphate deaminase (EC 3.5.4.17) is an enzyme that catalyzes the chemical reaction

Inosine-5'-monophosphate dehydrogenase 1, also known as IMP dehydrogenase 1, is an enzyme that in humans is encoded by the IMPDH1 gene.

Phosphoribosylglycinamide formyltransferase (EC 2.1.2.2), also known as glycinamide ribonucleotide transformylase (GAR Tfase), is an enzyme with systematic name 10-formyltetrahydrofolate:5'-phosphoribosylglycinamide N-formyltransferase. This enzyme catalyses the following chemical reaction
In molecular biology, the CBS domain is a protein domain found in a range of proteins in all species from bacteria to humans. It was first identified as a conserved sequence region in 1997 and named after cystathionine beta synthase, one of the proteins it is found in. CBS domains are also found in a wide variety of other proteins such as inosine monophosphate dehydrogenase, voltage gated chloride channels and AMP-activated protein kinase (AMPK). CBS domains regulate the activity of associated enzymatic and transporter domains in response to binding molecules with adenosyl groups such as AMP and ATP, or s-adenosylmethionine.

Inosine-5'-monophosphate dehydrogenase 2, also known as IMP dehydrogenase 2, is an enzyme that in humans is encoded by the IMPDH2 gene.
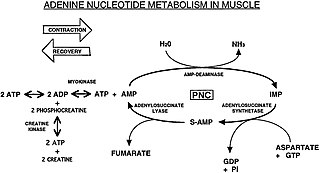
The Purine Nucleotide Cycle is a metabolic pathway in protein metabolism requiring the amino acids aspartate and glutamate. The cycle is used to regulate the levels of adenine nucleotides, in which ammonia and fumarate are generated. AMP converts into IMP and the byproduct ammonia. IMP converts to S-AMP (adenylosuccinate), which then converts to AMP and the byproduct fumarate. The fumarate goes on to produce ATP (energy) via oxidative phosphorylation as it enters the Krebs cycle and then the electron transport chain. Lowenstein first described this pathway and outlined its importance in processes including amino acid catabolism and regulation of flux through glycolysis and the Krebs cycle.
The gua operon is responsible for regulating the synthesis of guanosine mono phosphate (GMP), a purine nucleotide, from inosine monophosphate. It consists of two structural genes guaB (encodes for IMP dehydrogenase or and guaA apart from the promoter and operator region.




















