In biochemistry, denaturation is a process in which proteins or nucleic acids lose folded structure present in their native state due to various factors, including application of some external stress or compound, such as a strong acid or base, a concentrated inorganic salt, an organic solvent, agitation and radiation, or heat. If proteins in a living cell are denatured, this results in disruption of cell activity and possibly cell death. Protein denaturation is also a consequence of cell death. Denatured proteins can exhibit a wide range of characteristics, from conformational change and loss of solubility or dissociation of cofactors to aggregation due to the exposure of hydrophobic groups. The loss of solubility as a result of denaturation is called coagulation. Denatured proteins lose their 3D structure, and therefore, cannot function.

Proteolysis is the breakdown of proteins into smaller polypeptides or amino acids. Uncatalysed, the hydrolysis of peptide bonds is extremely slow, taking hundreds of years. Proteolysis is typically catalysed by cellular enzymes called proteases, but may also occur by intra-molecular digestion.

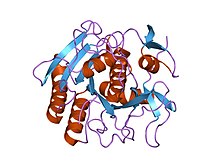
Trypsin is an enzyme in the first section of the small intestine that starts the digestion of protein molecules by cutting long chains of amino acids into smaller pieces. It is a serine protease from the PA clan superfamily, found in the digestive system of many vertebrates, where it hydrolyzes proteins. Trypsin is formed in the small intestine when its proenzyme form, the trypsinogen produced by the pancreas, is activated. Trypsin cuts peptide chains mainly at the carboxyl side of the amino acids lysine or arginine. It is used for numerous biotechnological processes. The process is commonly referred to as trypsinogen proteolysis or trypsinization, and proteins that have been digested/treated with trypsin are said to have been trypsinized.

A protease is an enzyme that catalyzes proteolysis, breaking down proteins into smaller polypeptides or single amino acids, and spurring the formation of new protein products. They do this by cleaving the peptide bonds within proteins by hydrolysis, a reaction where water breaks bonds. Proteases are involved in numerous biological pathways, including digestion of ingested proteins, protein catabolism, and cell signaling.
In biology and biochemistry, protease inhibitors, or antiproteases, are molecules that inhibit the function of proteases. Many naturally occurring protease inhibitors are proteins.

Serine proteases are enzymes that cleave peptide bonds in proteins. Serine serves as the nucleophilic amino acid at the (enzyme's) active site. They are found ubiquitously in both eukaryotes and prokaryotes. Serine proteases fall into two broad categories based on their structure: chymotrypsin-like (trypsin-like) or subtilisin-like.

Tosyl phenylalanyl chloromethyl ketone (TPCK) is a protease inhibitor. Its structural formula is 1-chloro-3-tosylamido-4-phenyl-2-butanone.

Papain, also known as papaya proteinase I, is a cysteine protease enzyme present in papaya and mountain papaya. It is the namesake member of the papain-like protease family.

Dithiothreitol (DTT) is an organosulfur compound with the formula (CH CH2SH)2. A colorless compound, it is classified as a dithiol and a diol. DTT is redox reagent also known as Cleland's reagent, after W. Wallace Cleland. The reagent is commonly used in its racemic form. Its name derives from the four-carbon sugar, threose. DTT has an epimeric ('sister') compound, dithioerythritol (DTE).

Cysteine proteases, also known as thiol proteases, are hydrolase enzymes that degrade proteins. These proteases share a common catalytic mechanism that involves a nucleophilic cysteine thiol in a catalytic triad or dyad.
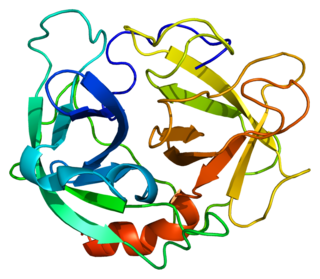
Neutrophil elastase is a serine proteinase in the same family as chymotrypsin and has broad substrate specificity. Neutrophil elastase is secreted by neutrophils during inflammation, and destroys bacteria and host tissue. It also localizes to neutrophil extracellular traps (NETs), via its high affinity for DNA, an unusual property for serine proteases.
Protein metabolism denotes the various biochemical processes responsible for the synthesis of proteins and amino acids (anabolism), and the breakdown of proteins by catabolism.

Thermolysin is a thermostable neutral metalloproteinase enzyme produced by the Gram-positive bacteria Bacillus thermoproteolyticus. It requires one zinc ion for enzyme activity and four calcium ions for structural stability. Thermolysin specifically catalyzes the hydrolysis of peptide bonds containing hydrophobic amino acids. However thermolysin is also widely used for peptide bond formation through the reverse reaction of hydrolysis. Thermolysin is the most stable member of a family of metalloproteinases produced by various Bacillus species. These enzymes are also termed 'neutral' proteinases or thermolysin -like proteinases (TLPs).

Cathepsin L1 is a protein that in humans is encoded by the CTSL1 gene. The protein is a cysteine cathepsin, a lysosomal cysteine protease that plays a major role in intracellular protein catabolism.
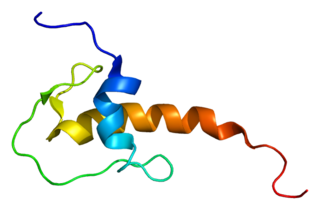
Lympho-epithelial Kazal-type-related inhibitor (LEKTI) also known as serine protease inhibitor Kazal-type 5 (SPINK5) is a protein that in humans is encoded by the SPINK5 gene.
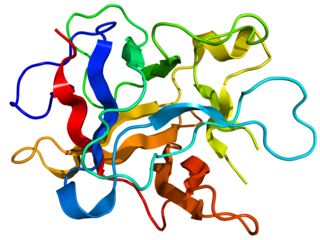
Kunitz soybean trypsin inhibitor is a type of protein contained in legume seeds which functions as a protease inhibitor. Kunitz-type Soybean Trypsin Inhibitors are usually specific for either trypsin or chymotrypsin. They are thought to protect seeds against consumption by animal predators.
Nepenthesin is an aspartic protease of plant origin that has so far been identified in the pitcher secretions of Nepenthes and in the leaves of Drosera peltata. It is similar to pepsin, but differs in that it also cleaves on either side of Asp residues and at Lys┼Arg. While more pH and temperature stable than porcine pepsin A, it is considerably less stable in urea or guanidine hydrochloride. It is the only known protein with such a stability profile.
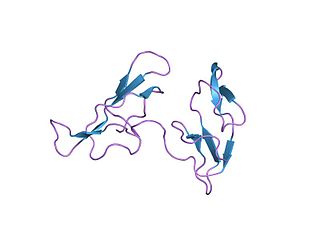
In molecular biology, the Bowman–Birk protease inhibitor family of proteins consists of eukaryotic proteinase inhibitors, belonging to MEROPS inhibitor family I12, clan IF. They mainly inhibit serine peptidases of the S1 family, but also inhibit S3 peptidases.
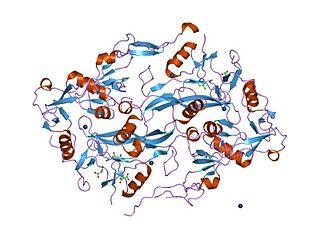
The Kazal domain is an evolutionary conserved protein domain usually indicative of serine protease inhibitors. However, kazal-like domains are also seen in the extracellular part of agrins, which are not known to be protease inhibitors.
Serratia marcescens nuclease is an enzyme. This enzyme catalyses the following chemical reaction