Severe acute respiratory syndrome–related coronavirus is a species of virus consisting of many known strains phylogenetically related to severe acute respiratory syndrome coronavirus 1 (SARS-CoV-1) that have been shown to possess the capability to infect humans, bats, and certain other mammals. These enveloped, positive-sense single-stranded RNA viruses enter host cells by binding to the angiotensin-converting enzyme 2 (ACE2) receptor. The SARSr-CoV species is a member of the genus Betacoronavirus and of the subgenus Sarbecovirus.

Vaccinia virus is a large, complex, enveloped virus belonging to the poxvirus family. It has a linear, double-stranded DNA genome approximately 190 kbp in length, which encodes approximately 250 genes. The dimensions of the virion are roughly 360 × 270 × 250 nm, with a mass of approximately 5–10 fg. The vaccinia virus is the source of the modern smallpox vaccine, which the World Health Organization (WHO) used to eradicate smallpox in a global vaccination campaign in 1958–1977. Although smallpox no longer exists in the wild, vaccinia virus is still studied widely by scientists as a tool for gene therapy and genetic engineering.
Polyadenylation is the addition of a poly(A) tail to an RNA transcript, typically a messenger RNA (mRNA). The poly(A) tail consists of multiple adenosine monophosphates; in other words, it is a stretch of RNA that has only adenine bases. In eukaryotes, polyadenylation is part of the process that produces mature mRNA for translation. In many bacteria, the poly(A) tail promotes degradation of the mRNA. It, therefore, forms part of the larger process of gene expression.
In molecular biology, the five-prime cap is a specially altered nucleotide on the 5′ end of some primary transcripts such as precursor messenger RNA. This process, known as mRNA capping, is highly regulated and vital in the creation of stable and mature messenger RNA able to undergo translation during protein synthesis. Mitochondrial mRNA and chloroplastic mRNA are not capped.
In cellular biology, P-bodies, or processing bodies, are distinct foci formed by phase separation within the cytoplasm of a eukaryotic cell consisting of many enzymes involved in mRNA turnover. P-bodies are highly conserved structures and have been observed in somatic cells originating from vertebrates and invertebrates, plants and yeast. To date, P-bodies have been demonstrated to play fundamental roles in general mRNA decay, nonsense-mediated mRNA decay, adenylate-uridylate-rich element mediated mRNA decay, and microRNA (miRNA) induced mRNA silencing. Not all mRNAs which enter P-bodies are degraded, as it has been demonstrated that some mRNAs can exit P-bodies and re-initiate translation. Purification and sequencing of the mRNA from purified processing bodies showed that these mRNAs are largely translationally repressed upstream of translation initiation and are protected from 5' mRNA decay.

A viroplasm, sometimes called "virus factory" or "virus inclusion", is an inclusion body in a cell where viral replication and assembly occurs. They may be thought of as viral factories in the cell. There are many viroplasms in one infected cell, where they appear dense to electron microscopy. Very little is understood about the mechanism of viroplasm formation.

5′-3′ exoribonuclease 1 (Xrn1) is a protein that in humans is encoded by the XRN1 gene. Xrn1 hydrolyses RNA in the 5′ to 3′ direction.
In enzymology, a mRNA (guanine-N7-)-methyltransferase also known as mRNA cap guanine-N7 methyltransferase is an enzyme that catalyzes the chemical reaction

Ras GTPase-activating protein-binding protein 1 is an enzyme that in humans is encoded by the G3BP1 gene.

Lysyl-tRNA synthetase is an enzyme that in humans is encoded by the KARS gene.

Trinucleotide repeat-containing gene 6A protein is a protein that in humans is encoded by the TNRC6A gene.

mRNA-decapping enzyme 2 is a protein that in humans is encoded by the DCP2 gene.

Scavenger mRNA-decapping enzyme DcpS is a protein that in humans is encoded by the DCPS gene.

mRNA-decapping enzyme 1A is a protein that in humans is encoded by the DCP1A gene.

Caprin-1 is a protein that in humans is encoded by the CAPRIN1 gene. It is suggested that Caprin1 is essential for the formation of long-term memory.

mRNA-decapping enzyme 1B is a protein that in humans is encoded by the DCP1B gene.
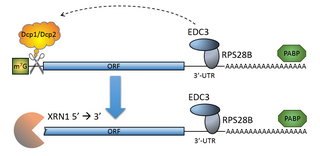
The process of messenger RNA decapping consists of hydrolysis of the 5' cap structure on the RNA exposing a 5' monophosphate. In eukaryotes, this 5' monophosphate is a substrate for the 5' exonuclease Xrn1 and the mRNA is quickly destroyed. There are many situations which may lead to the removal of the cap, some of which are discussed below.

The mRNA decapping complex is a protein complex in eukaryotic cells responsible for removal of the 5' cap. The active enzyme of the decapping complex is the bilobed Nudix family enzyme Dcp2, which hydrolyzes 5' cap and releases 7mGDP and a 5'-monophosphorylated mRNA. This decapped mRNA is inhibited for translation and will be degraded by exonucleases. The core decapping complex is conserved in eukaryotes. Dcp2 is activated by Decapping Protein 1 (Dcp1) and in higher eukaryotes joined by the scaffold protein VCS. Together with many other accessory proteins, the decapping complex assembles in P-bodies in the cytoplasm.
M7GpppX diphosphatase (EC 3.6.1.59, DcpS, m7GpppX pyrophosphatase, m7GpppN m7GMP phosphohydrolase) is an enzyme with systematic name m7G5'ppp5'N m7GMP phosphohydrolase. This enzyme catalyses the following chemical reaction

MicroRNA 141 is a non-coding RNA molecule that in humans is encoded by the MIR141 gene.