Catechol dioxygenases are metalloprotein enzymes that carry out the oxidative cleavage of catechols. This class of enzymes incorporate dioxygen into the substrate. Catechol dioxygenases belong to the class of oxidoreductases and have several different substrate specificities, including catechol 1,2-dioxygenase, catechol 2,3-dioxygenase, and protocatechuate 3,4-dioxygenase. The active site of catechol dioxygenases most frequently contains iron, but manganese-containing forms are also known.

Muconate lactonizing enzymes are involved in the breakdown of lignin-derived aromatics, catechol and protocatechuate, to citric acid cycle intermediates as a part of the β-ketoadipate pathway in soil microbes. Some bacterial species are also capable of dehalogenating chloroaromatic compounds by the action of chloromuconate lactonizing enzymes. MLEs consist of several strands which have variable reaction favorable parts therefore the configuration of the strands affect its ability to accept protons. The bacterial MLEs belong to the enolase superfamily, several structures from which are known. MLEs have an identifying structure made up of two proteins and two Magnesium ions as well as various classes depending on whether it is bacterial or eukaryotic. The reaction mechanism that MLEs undergo are the reverse of beta-elimination in which the enolate alpha-carbon is protonated. MLEs can undergo mutations caused by a deletion of catB structural genes which can cause some bacteria to lose its functions such as the ability to grow. Additional mutations to MLEs can cause its structure and function to alter and could cause the conformation to change therefore making it an inactive enzyme that is unable to bind its substrate. There is another enzyme called Mandelate Racemase that is very similar to MLEs in the structural way as well as them both being a part of the enolase superfamily. They both have the same end product even though they undergo different chemical reactions in order to reach the end product.

Catechol 1,2- dioxygenase is an enzyme that catalyzes the oxidative ring cleavage of catechol to form cis,cis-muconic acid:
In enzymology, a 2-oxoadipate reductase (EC 1.1.1.172) is an enzyme that catalyzes the chemical reaction
In enzymology, a 3-carboxy-cis,cis-muconate cycloisomerase is an enzyme that catalyzes the chemical reaction
In enzymology, a muconolactone Δ-isomerase is an enzyme that catalyzes the chemical reaction
The enzyme 3-chloro-D-alanine dehydrochlorinase (EC 4.5.1.2) catalyzes the reaction
The enzyme 4-carboxymuconolactone decarboxylase (EC 4.1.1.44) catalyzes the chemical reaction
The enzyme 1,4-lactonase (EC 3.1.1.25) catalyzes the generic reaction
The enzyme 2-pyrone-4,6-dicarboxylate lactonase (EC 3.1.1.57, LigI) catalyzes the reversible hydrolytic reaction
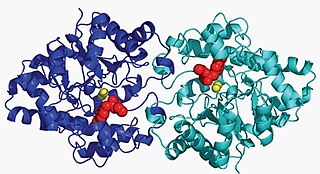
Aryldialkylphosphatase is a metalloenzyme that hydrolyzes the triester linkage found in organophosphate insecticides:
In enzymology, a carboxymethylenebutenolidase (EC 3.1.1.45, also known as CMBL and dienelactone hydrolase) is an enzyme that catalyzes the chemical reaction
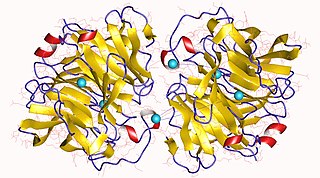
The enzyme gluconolactonase (EC 3.1.1.17) catalyzes the reaction
The enzyme limonin-D-ring-lactonase (EC 3.1.1.36) catalyzes the reaction
The enzyme L-rhamnono-1,4-lactonase (EC 3.1.1.65) catalyzes the reaction
The enzyme xylono-1,4-lactonase (EC 3.1.1.68) catalyzes the reaction
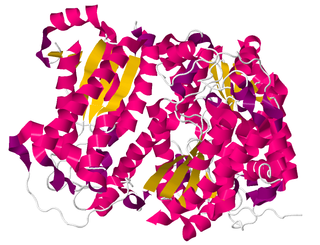
In enzymology, a creatininase (EC 3.5.2.10) is an enzyme that catalyses the hydrolysis of creatinine to creatine, which can then be metabolised to urea and sarcosine by creatinase.

The alpha/beta hydrolase superfamily is a superfamily of hydrolytic enzymes of widely differing phylogenetic origin and catalytic function that share a common fold. The core of each enzyme is an alpha/beta-sheet, containing 8 beta strands connected by 6 alpha helices. The enzymes are believed to have diverged from a common ancestor, retaining little obvious sequence similarity, but preserving the arrangement of the catalytic residues. All have a catalytic triad, the elements of which are borne on loops, which are the best-conserved structural features of the fold.

Lactonase (EC 3.1.1.81, acyl-homoserine lactonase; systematic name N-acyl-L-homoserine-lactone lactonohydrolase) is a metalloenzyme, produced by certain species of bacteria, which targets and inactivates acylated homoserine lactones (AHLs). It catalyzes the reaction
2-Hydroxymuconate-6-semialdehyde dehydrogenase (EC 1.2.1.85, xylG [gene], praB [gene] ) is an enzyme with systematic name (2E,4Z)-2-hydroxy-6-oxohexa-2,4-dienoate:NAD+ oxidoreductase. This enzyme catalyses the following chemical reaction








