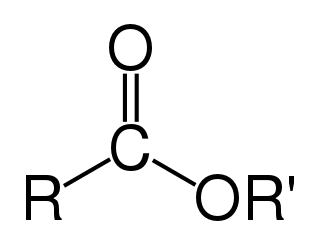
In chemistry, an ester is a functional group derived from an acid in which the hydrogen atom (H) of at least one acidic hydroxyl group of that acid is replaced by an organyl group. Analogues derived from oxygen replaced by other chalcogens belong to the ester category as well. According to some authors, organyl derivatives of acidic hydrogen of other acids are esters as well, but not according to the IUPAC.

An acetate is a salt formed by the combination of acetic acid with a base. "Acetate" also describes the conjugate base or ion typically found in aqueous solution and written with the chemical formula C
2H
3O−
2. The neutral molecules formed by the combination of the acetate ion and a positive ion are also commonly called "acetates". The simplest of these is hydrogen acetate with corresponding salts, esters, and the polyatomic anion CH
3CO−
2, or CH
3COO−
.
In biochemistry, hydrolases constitute a class of enzymes that commonly function as biochemical catalysts that use water to break a chemical bond:

Acetylacetone is an organic compound with the chemical formula CH3−C(=O)−CH2−C(=O)−CH3. It is classified as a 1,3-diketone. It exists in equilibrium with a tautomer CH3−C(=O)−CH=C(−OH)−CH3. The mixture is a colorless liquid. These tautomers interconvert so rapidly under most conditions that they are treated as a single compound in most applications. Acetylacetone is a building block for the synthesis of many coordination complexes as well as heterocyclic compounds.

An organic acid anhydride is an acid anhydride that is also an organic compound. An acid anhydride is a compound that has two acyl groups bonded to the same oxygen atom. A common type of organic acid anhydride is a carboxylic anhydride, where the parent acid is a carboxylic acid, the formula of the anhydride being (RC(O))2O. Symmetrical acid anhydrides of this type are named by replacing the word acid in the name of the parent carboxylic acid by the word anhydride. Thus, (CH3CO)2O is called acetic anhydride.Mixed (or unsymmetrical) acid anhydrides, such as acetic formic anhydride (see below), are known, whereby reaction occurs between two different carboxylic acids. Nomenclature of unsymmetrical acid anhydrides list the names of both of the reacted carboxylic acids before the word "anhydride" (for example, the dehydration reaction between benzoic acid and propanoic acid would yield "benzoic propanoic anhydride").
In biochemistry, sulfatases EC 3.1.6.- are a class of enzymes of the esterase class that catalyze the hydrolysis of sulfate esters into an alcohol and a bisulfate:
In chemistry, carbonylation refers to reactions that introduce carbon monoxide (CO) into organic and inorganic substrates. Carbon monoxide is abundantly available and conveniently reactive, so it is widely used as a reactant in industrial chemistry. The term carbonylation also refers to oxidation of protein side chains.
Aryldialkylphosphatase is a metalloenzyme that hydrolyzes the triester linkage found in organophosphate insecticides:
The enzyme arylesterase (EC 3.1.1.2) catalyzes the reaction
The enzyme carboxylesterase (or carboxylic-ester hydrolase, EC 3.1.1.1; systematic name carboxylic-ester hydrolase) catalyzes reactions of the following form:
The enzyme cephalosporin-C deacetylase (EC 3.1.1.41) catalyzes the reaction
The enzyme sialate O-acetylesterase (EC 3.1.1.53) catalyzes the reaction
Liver carboxylesterase 1 also known as carboxylesterase 1 is an enzyme that in humans is encoded by the CES1 gene. The protein is also historically known as serine esterase 1 (SES1), monocyte esterase and cholesterol ester hydrolase (CEH). Three transcript variants encoding three different isoforms have been found for this gene. The various protein products from isoform a, b and c range in size from 568, 567 and 566 amino acids long, respectively.

Chromosome 11 open reading frame 54 (C11orf54) is a protein that in humans is encoded by the C11orf54 gene. The "Homo sapiens" gene, C11orf54 is also known as PTD012 and PTOD12. C11orf54 exhibits hydrolase activity on p-nitrophenyl acetate and acts on ester bonds, though the overall function is still not fully understood by the scientific community. The protein is highly conserved with the most distant homolog found is in bacteria.

Acetic acid, systematically named ethanoic acid, is an acidic, colourless liquid and organic compound with the chemical formula CH3COOH. Vinegar is at least 4% acetic acid by volume, making acetic acid the main component of vinegar apart from water. It has been used, as a component of vinegar, throughout history from at least the third century BC. Acetic acid is also known as acetyl hydroxide (AcOH).
CAZy is a database of Carbohydrate-Active enZYmes (CAZymes). The database contains a classification and associated information about enzymes involved in the synthesis, metabolism, and recognition of complex carbohydrates, i.e. disaccharides, oligosaccharides, polysaccharides, and glycoconjugates. Included in the database are families of glycoside hydrolases, glycosyltransferases, polysaccharide lyases, carbohydrate esterases, and non-catalytic carbohydrate-binding modules. The CAZy database also includes a classification of Auxiliary Activity redox enzymes involved in the breakdown of lignocellulose.

The enzyme triacylglycerol lipase (also triglyceride lipase, EC 3.1.1.3;systematic name triacylglycerol acylhydrolase) catalyses the hydrolysis of ester linkages of triglycerides:
The enzyme acetylajmaline esterase (EC 3.1.1.80, AAE, 2β(R)-17-O-acetylajmalan:acetylesterase, acetylajmalan esterase; systematic name 17-O-acetylajmaline O-acetylhydrolase) catalyses the following reactions:
The enzyme pyrethroid hydrolase (EC 3.1.1.88, pyrethroid-hydrolyzing carboxylesterase, pyrethroid-hydrolysing esterase, pyrethroid-hydrolyzing esterase, pyrethroid-selective esterase, pyrethroid-cleaving enzyme, permethrinase, PytH, EstP; systematic name pyrethroid-ester hydrolase) catalyses the reaction
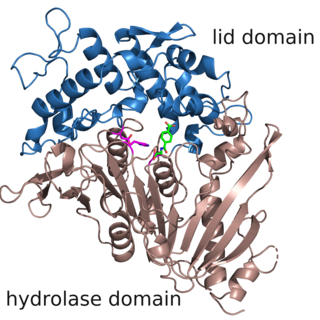
The enzyme MHETase is a hydrolase, which was discovered in 2016. It cleaves 2-hydroxyethyl terephthalic acid, the PET degradation product by PETase, to ethylene glycol and terephthalic acid. This pair of enzymes, PETase and MHETase, enable the bacterium Ideonella sakaiensis to live on the plastic PET as sole carbon source.











