Hydrolase is a class of enzymes that commonly perform as biochemical catalysts that use water to break a chemical bond, which typically results in dividing a larger molecule into smaller molecules. Some common examples of hydrolase enzymes are esterases including lipases, phosphatases, glycosidases, peptidases, and nucleosidases.
The enzyme 1-alkyl-2-acetylglycerophosphocholine esterase (EC 3.1.1.47) catalyzes the reaction
The enzyme 5-(3,4-diacetoxybut-1-ynyl)-2,2′-bithiophene deacetylase (EC 3.1.1.66) catalyzes the reaction
The enzyme acetoxybutynylbithiophene deacetylase (EC 3.1.1.54) catalyzes the reaction
The enzyme acetyl-CoA hydrolase catalyzes the reaction
The enzyme acetylesterase (EC 3.1.1.6) catalyzes the reaction
The enzyme acetylsalicylate deacetylase (EC 3.1.1.55) catalyzes the reaction
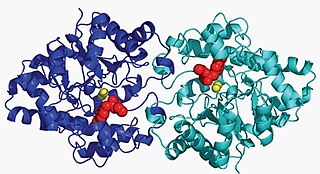
Aryldialkylphosphatase is a metalloenzyme that hydrolyzes the triester linkage found in organophosphate insecticides:
The enzyme arylesterase (EC 3.1.1.2) catalyzes the reaction
The enzyme carboxylesterase (or carboxylic-ester hydrolase, EC 3.1.1.1; systematic name carboxylic-ester hydrolase) catalyzes reactions of the following form:
The enzyme cephalosporin-C deacetylase (EC 3.1.1.41) catalyzes the reaction

The enzyme juvenile hormone esterase (EC 3.1.1.59, systematic name methyl-(2E,6E,10R)-10,11-epoxy-3,7,11-trimethyltrideca-2,6-dienoate acylhydrolase, JH esterase) catalyzes the hydrolysis of juvenile hormone:
The enzyme lysophospholipase (EC 3.1.1.5) catalyzes the reaction
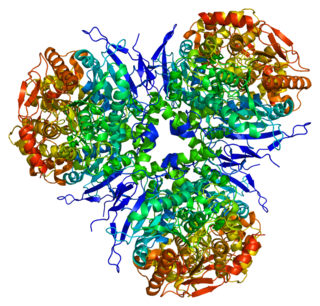
Liver carboxylesterase 1 also known as carboxylesterase 1 is an enzyme that in humans is encoded by the CES1 gene. The protein is also historically known as serine esterase 1 (SES1), monocyte esterase and cholesterol ester hydrolase (CEH). Three transcript variants encoding three different isoforms have been found for this gene. The various protein products from isoform a, b and c range in size from 568, 567 and 566 amino acids long, respectively.

Neuropathy target esterase, also known as patatin-like phospholipase domain-containing protein 6 (PNPLA6), is an esterase enzyme that in humans is encoded by the PNPLA6 gene.
The enzyme acetylajmaline esterase (EC 3.1.1.80, AAE, 2β(R)-17-O-acetylajmalan:acetylesterase, acetylajmalan esterase; systematic name 17-O-acetylajmaline O-acetylhydrolase) catalyses the following reactions:

Jagannath Ganguly (1921–2007) was an Indian biochemist known for his researches on Vitamin A and fatty acids, which assisted in the better understanding of their metabolism in humans. Born on the 1 April 1921, he authored a book, Biochemistry of Vitamin A, which details the physiological, biochemical and nutritional characteristics of the organic compound. The Council of Scientific and Industrial Research, the apex agency of the Government of India for scientific research, awarded him the Shanti Swarup Bhatnagar Prize for Science and Technology, one of the highest Indian science awards, in 1963, for his contributions to biological sciences. He died on 12 December 2007.
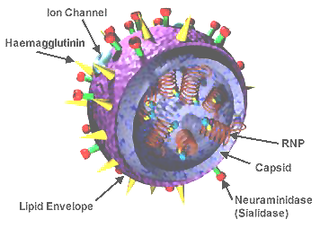
Influenza D virus is a species in the virus genus Deltainfluenzavirus, in the family Orthomyxoviridae, that causes influenza.
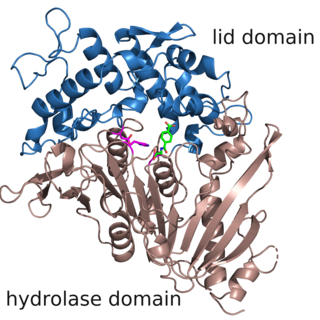
The enzyme MHETase is a hydrolase, which was discovered in 2016. It cleaves 2-hydroxyethyl terephthalic acid, the PET degradation product by PETase, to ethylene glycol and terephthalic acid. This pair of enzymes, PETase and MHETase, enable the bacterium Ideonella sakaiensis to live on the plastic PET as sole carbon source.







