Related Research Articles

A protease is an enzyme that catalyzes proteolysis, breaking down proteins into smaller polypeptides or single amino acids, and spurring the formation of new protein products. They do this by cleaving the peptide bonds within proteins by hydrolysis, a reaction where water breaks bonds. Proteases are involved in numerous biological pathways, including digestion of ingested proteins, protein catabolism, and cell signaling.

Ranpirnase is a ribonuclease enzyme found in the oocytes of the Northern Leopard Frog. Ranpirnase is a member of the pancreatic ribonuclease protein superfamily and degrades RNA substrates with a sequence preference for uracil and guanine nucleotides. Along with amphinase, another leopard frog ribonuclease, Ranpirnase has been studied as a potential cancer and antiviral treatment due to its unusual mechanism of cytotoxicity tested against transformed cells and antiviral activity.
Nitrilase enzymes catalyse the hydrolysis of nitriles to carboxylic acids and ammonia, without the formation of "free" amide intermediates. Nitrilases are involved in natural product biosynthesis and post translational modifications in plants, animals, fungi and certain prokaryotes. Nitrilases can also be used as catalysts in preparative organic chemistry. Among others, nitrilases have been used for the resolution of racemic mixtures. Nitrilase should not be confused with nitrile hydratase which hydrolyses nitriles to amides. Nitrile hydratases are almost invariably co-expressed with an amidase, which converts the amide to the carboxylic acid. Consequently, it can sometimes be difficult to distinguish nitrilase activity from nitrile hydratase plus amidase activity.

Muconate lactonizing enzymes are involved in the breakdown of lignin-derived aromatics, catechol and protocatechuate, to citric acid cycle intermediates as a part of the β-ketoadipate pathway in soil microbes. Some bacterial species are also capable of dehalogenating chloroaromatic compounds by the action of chloromuconate lactonizing enzymes. MLEs consist of several strands which have variable reaction favorable parts therefore the configuration of the strands affect its ability to accept protons. The bacterial MLEs belong to the enolase superfamily, several structures from which are known. MLEs have an identifying structure made up of two proteins and two Magnesium ions as well as various classes depending on whether it is bacterial or eukaryotic. The reaction mechanism that MLEs undergo are the reverse of beta-elimination in which the enolate alpha-carbon is protonated. MLEs can undergo mutations caused by a deletion of catB structural genes which can cause some bacteria to lose its functions such as the ability to grow. Additional mutations to MLEs can cause its structure and function to alter and could cause the conformation to change therefore making it an inactive enzyme that is unable to bind its substrate. There is another enzyme called Mandelate Racemase that is very similar to MLEs in the structural way as well as them both being a part of the enolase superfamily. They both have the same end product even though they undergo different chemical reactions in order to reach the end product.
Phospholipase D (EC 3.1.4.4, lipophosphodiesterase II, lecithinase D, choline phosphatase, PLD; systematic name phosphatidylcholine phosphatidohydrolase) is an enzyme of the phospholipase superfamily that catalyses the following reaction
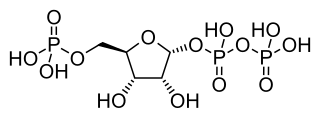
Phosphoribosyl pyrophosphate (PRPP) is a pentose phosphate. It is a biochemical intermediate in the formation of purine nucleotides via inosine-5-monophosphate, as well as in pyrimidine nucleotide formation. Hence it is a building block for DNA and RNA. The vitamins thiamine and cobalamin, and the amino acid tryptophan also contain fragments derived from PRPP. It is formed from ribose 5-phosphate (R5P) by the enzyme ribose-phosphate diphosphokinase:
The MetaCyc database is one of the largest metabolic pathways and enzymes databases currently available. The data in the database is manually curated from the scientific literature, and covers all domains of life. MetaCyc has extensive information about chemical compounds, reactions, metabolic pathways and enzymes. The data have been curated from more than 58,000 publications.
Barbiturase is a zinc-containing amidohydrolase. Its systemic name is barbiturate amidohydrolase (3-oxo-3-ureidopropanoate-forming). Barbiturase acts as a catalyst in the second step of oxidative pyrimidine degradation, promoting the ring-opening hydrolysis of barbituric acid to ureidomalonic acid. Although grouped into the naturally existing amidohydrolases, it demonstrates more homology with cyanuric acid amidohydrolase. Therefore, it has been proposed that barbiturase, along with cyanuric acid, should be grouped into a new family. KEGG
In enzymology, a manganese peroxidase (EC 1.11.1.13) is an enzyme that catalyzes the chemical reaction
In enzymology, a muconolactone Δ-isomerase is an enzyme that catalyzes the chemical reaction

The enzyme 4-hydroxy-2-oxovalerate aldolase catalyzes the chemical reaction
The enzyme 4-hydroxy-4-methyl-2-oxoglutarate aldolase catalyzes the chemical reaction

In enzymology, N-acetylglucosamine-6-phosphate deacetylase (EC 3.5.1.25), also known as GlcNAc-6-phosphate deacetylase or NagA, is an enzyme that catalyzes the deacetylation of N-acetylglucosamine-6-phosphate (GlcNAc-6-P) to glucosamine-6-phosphate (GlcN-6-P):

Fatty-acyl-CoA synthase, or more commonly known as yeast fatty acid synthase, is an enzyme complex responsible for fatty acid biosynthesis, and is of Type I Fatty Acid Synthesis (FAS). Yeast fatty acid synthase plays a pivotal role in fatty acid synthesis. It is a 2.6 MDa barrel shaped complex and is composed of two, unique multi-functional subunits: alpha and beta. Together, the alpha and beta units are arranged in an α6β6 structure. The catalytic activities of this enzyme complex involves a coordination system of enzymatic reactions between the alpha and beta subunits. The enzyme complex therefore consists of six functional centers for fatty acid synthesis.
Dioxygenases are oxidoreductase enzymes. Aerobic life, from simple single-celled bacteria species to complex eukaryotic organisms, has evolved to depend on the oxidizing power of dioxygen in various metabolic pathways. From energetic adenosine triphosphate (ATP) generation to xenobiotic degradation, the use of dioxygen as a biological oxidant is widespread and varied in the exact mechanism of its use. Enzymes employ many different schemes to use dioxygen, and this largely depends on the substrate and reaction at hand.
2-hydroxy-4-carboxymuconate semialdehyde hemiacetal dehydrogenase (EC 1.1.1.312, 2-hydroxy-4-carboxymuconate 6-semialdehyde dehydrogenase, 4-carboxy-2-hydroxy-cis,cis-muconate-6-semialdehyde:NADP+ oxidoreductase, alpha-hydroxy-gamma-carboxymuconic epsilon-semialdehyde dehydrogenase, 4-carboxy-2-hydroxymuconate-6-semialdehyde dehydrogenase, LigC, ProD) is an enzyme with systematic name 4-carboxy-2-hydroxymuconate semialdehyde hemiacetal:NADP+ 2-oxidoreductase. This enzyme catalyses the following chemical reaction
2-Hydroxymuconate-6-semialdehyde dehydrogenase (EC 1.2.1.85, xylG [gene], praB [gene] ) is an enzyme with systematic name (2E,4Z)-2-hydroxy-6-oxohexa-2,4-dienoate:NAD+ oxidoreductase. This enzyme catalyses the following chemical reaction
2-hydroxy-6-oxo-6-(2-aminophenyl)hexa-2,4-dienoate hydrolase (EC 3.7.1.13, CarC) is an enzyme with systematic name (2E,4E)-6-(2-aminophenyl)-2-hydroxy-6-oxohexa-2,4-dienoate acylhydrolase. This enzyme catalyses the following chemical reaction
2-hydroxymuconate tautomerase is an enzyme with systematic name (2Z,4E)-2-hydroxyhexa-2,4-dienedioate keto-enol isomerase. This enzyme catalyses the following chemical reaction

6-Amyl-α-pyrone, also 6-pentyl-2-pyrone or 6PP, is an unsaturated lactone molecule. It contains two double bonds in the ring and a pentyl substituent at carbon adjacent to the ring oxygen. It is a colorless liquid which possesses characteristic coconut aroma, produced biologically by Trichoderma species. It is found in animal foods, peach, and heated beef.
References
- ↑ "syringate degradation". MetaCyc. SRI International.
- ↑ "protocatechuate degradation I (meta-cleavage pathway)". MetaCyc. SRI International.
- 1 2 Hobbs ME, Malashkevich V, Williams HJ, Xu C, Sauder JM, Burley SK, Almo SC, Raushel FM (April 2012). "Structure and Catalytic Mechanism of LigI: Insight into the Amidohydrolase Enzymes of cog3618 and Lignin Degradation". Biochemistry. 51 (16): 3497–507. doi:10.1021/bi300307b. PMC 3416963 . PMID 22475079.