Hydrolysis is any chemical reaction in which a molecule of water breaks one or more chemical bonds. The term is used broadly for substitution, elimination, and solvation reactions in which water is the nucleophile.
A protein phosphatase is a phosphatase enzyme that removes a phosphate group from the phosphorylated amino acid residue of its substrate protein. Protein phosphorylation is one of the most common forms of reversible protein posttranslational modification (PTM), with up to 30% of all proteins being phosphorylated at any given time. Protein kinases (PKs) are the effectors of phosphorylation and catalyse the transfer of a γ-phosphate from ATP to specific amino acids on proteins. Several hundred PKs exist in mammals and are classified into distinct super-families. Proteins are phosphorylated predominantly on Ser, Thr and Tyr residues, which account for 79.3, 16.9 and 3.8% respectively of the phosphoproteome, at least in mammals. In contrast, protein phosphatases (PPs) are the primary effectors of dephosphorylation and can be grouped into three main classes based on sequence, structure and catalytic function. The largest class of PPs is the phosphoprotein phosphatase (PPP) family comprising PP1, PP2A, PP2B, PP4, PP5, PP6 and PP7, and the protein phosphatase Mg2+- or Mn2+-dependent (PPM) family, composed primarily of PP2C. The protein Tyr phosphatase (PTP) super-family forms the second group, and the aspartate-based protein phosphatases the third. The protein pseudophosphatases form part of the larger phosphatase family, and in most cases are thought to be catalytically inert, instead functioning as phosphate-binding proteins, integrators of signalling or subcellular traps. Examples of membrane-spanning protein phosphatases containing both active (phosphatase) and inactive (pseudophosphatase) domains linked in tandem are known, conceptually similar to the kinase and pseudokinase domain polypeptide structure of the JAK pseudokinases. A complete comparative analysis of human phosphatases and pseudophosphatases has been completed by Manning and colleagues, forming a companion piece to the ground-breaking analysis of the human kinome, which encodes the complete set of ~536 human protein kinases.
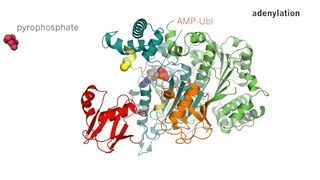
Enzyme catalysis is the increase in the rate of a process by a biological molecule, an "enzyme". Most enzymes are proteins, and most such processes are chemical reactions. Within the enzyme, generally catalysis occurs at a localized site, called the active site.
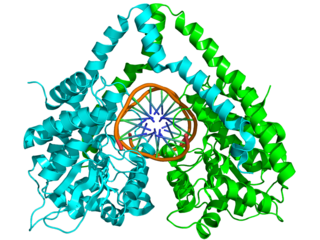
HindIII (pronounced "Hin D Three") is a type II site-specific deoxyribonuclease restriction enzyme isolated from Haemophilus influenzae that cleaves the DNA palindromic sequence AAGCTT in the presence of the cofactor Mg2+ via hydrolysis.

Cyclic ADP Ribose, frequently abbreviated as cADPR, is a cyclic adenine nucleotide (like cAMP) with two phosphate groups present on 5' OH of the adenosine (like ADP), further connected to another ribose at the 5' position, which, in turn, closes the cycle by glycosidic bonding to the nitrogen 1 (N1) of the same adenine base (whose position N9 has the glycosidic bond to the other ribose). The N1-glycosidic bond to adenine is what distinguishes cADPR from ADP-ribose (ADPR), the non-cyclic analog. cADPR is produced from nicotinamide adenine dinucleotide (NAD+) by ADP-ribosyl cyclases (EC 3.2.2.5) as part of a second messenger system.
Transition state analogs, are chemical compounds with a chemical structure that resembles the transition state of a substrate molecule in an enzyme-catalyzed chemical reaction. Enzymes interact with a substrate by means of strain or distortions, moving the substrate towards the transition state. Transition state analogs can be used as inhibitors in enzyme-catalyzed reactions by blocking the active site of the enzyme. Theory suggests that enzyme inhibitors which resembled the transition state structure would bind more tightly to the enzyme than the actual substrate. Examples of drugs that are transition state analog inhibitors include flu medications such as the neuraminidase inhibitor oseltamivir and the HIV protease inhibitors saquinavir in the treatment of AIDS.

Inorganic pyrophosphatase is an enzyme that catalyzes the conversion of one ion of pyrophosphate to two phosphate ions. This is a highly exergonic reaction, and therefore can be coupled to unfavorable biochemical transformations in order to drive these transformations to completion. The functionality of this enzyme plays a critical role in lipid metabolism, calcium absorption and bone formation, and DNA synthesis, as well as other biochemical transformations.

6-Phosphogluconolactonase (EC 3.1.1.31, 6PGL, PGLS, systematic name 6-phospho-D-glucono-1,5-lactone lactonohydrolase) is a cytosolic enzyme found in all organisms that catalyzes the hydrolysis of 6-phosphogluconolactone to 6-phosphogluconic acid in the oxidative phase of the pentose phosphate pathway:

Sucrose phosphorylase is an important enzyme in the metabolism of sucrose and regulation of other metabolic intermediates. Sucrose phosphorylase is in the class of hexosyltransferases. More specifically it has been placed in the retaining glycoside hydrolases family although it catalyzes a transglycosidation rather than hydrolysis. Sucrose phosphorylase catalyzes the conversion of sucrose to D-fructose and α-D-glucose-1-phosphate. It has been shown in multiple experiments that the enzyme catalyzes this conversion by a double displacement mechanism.
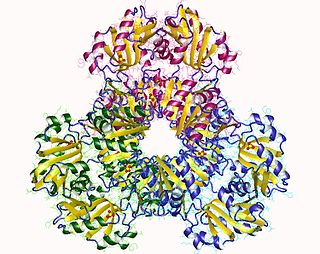
Ribose-phosphate diphosphokinase is an enzyme that converts ribose 5-phosphate into phosphoribosyl pyrophosphate (PRPP). It is classified under EC 2.7.6.1.

ADP-ribosylation is the addition of one or more ADP-ribose moieties to a protein. It is a reversible post-translational modification that is involved in many cellular processes, including cell signaling, DNA repair, gene regulation and apoptosis. Improper ADP-ribosylation has been implicated in some forms of cancer. It is also the basis for the toxicity of bacterial compounds such as cholera toxin, diphtheria toxin, and others.
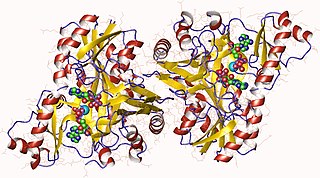
In enzymology, a biotin carboxylase (EC 6.3.4.14) is an enzyme that catalyzes the chemical reaction
In enzymology, an ADP-sugar diphosphatase (EC 3.6.1.21) is an enzyme that catalyzes the chemical reaction
In enzymology, an ATP diphosphatase (EC 3.6.1.8) is an enzyme that catalyzes the chemical reaction

In enzymology, a nucleoside-diphosphatase (EC 3.6.1.6) is an enzyme that catalyzes the chemical reaction

ADP-ribose pyrophosphatase, mitochondrial is an enzyme that in humans is encoded by the NUDT9 gene.
NUDIX hydrolases are a superfamily of hydrolytic enzymes capable of cleaving nucleoside diphosphates linked to x, hence their name. The reaction yields nucleoside monophosphate (NMP) plus X-P. Substrates hydrolysed by nudix enzymes comprise a wide range of organic pyrophosphates, including nucleoside di- and triphosphates, dinucleoside and diphosphoinositol polyphosphates, nucleotide sugars and RNA caps, with varying degrees of substrate specificity. Enzymes of the NUDIX superfamily are found in all types of organisms, including eukaryotes, bacteria and archaea.

In molecular biology, the (ADP-ribosyl)hydrolase (ARH) family contains enzymes which catalyses the hydrolysis of ADP-ribosyl modifications from proteins, nucleic acids and small molecules.

Nucleotide pyrophosphatase/phosphodiesterase (NPP) is a class of dimeric enzymes that catalyze the hydrolysis of phosphate diester bonds. NPP belongs to the alkaline phosphatase (AP) superfamily of enzymes. Humans express seven known NPP isoforms, some of which prefer nucleotide substrates, some of which prefer phospholipid substrates, and others of which prefer substrates that have not yet been determined. In eukaryotes, most NPPs are located in the cell membrane and hydrolyze extracellular phosphate diesters to affect a wide variety of biological processes. Bacterial NPP is thought to localize to the periplasm.
Mn2+-dependent ADP-ribose/CDP-alcohol diphosphatase (EC 3.6.1.53, Mn2+-dependent ADP-ribose/CDP-alcohol pyrophosphatase, ADPRibase-Mn) is an enzyme with systematic name CDP-choline phosphohydrolase. This enzyme catalyses the following chemical reaction

















