A restriction enzyme, restriction endonuclease, REase, ENase orrestrictase is an enzyme that cleaves DNA into fragments at or near specific recognition sites within molecules known as restriction sites. Restriction enzymes are one class of the broader endonuclease group of enzymes. Restriction enzymes are commonly classified into five types, which differ in their structure and whether they cut their DNA substrate at their recognition site, or if the recognition and cleavage sites are separate from one another. To cut DNA, all restriction enzymes make two incisions, once through each sugar-phosphate backbone of the DNA double helix.
In molecular biology, endonucleases are enzymes that cleave the phosphodiester bond within a polynucleotide chain. Some, such as deoxyribonuclease I, cut DNA relatively nonspecifically, while many, typically called restriction endonucleases or restriction enzymes, cleave only at very specific nucleotide sequences. Endonucleases differ from exonucleases, which cleave the ends of recognition sequences instead of the middle (endo) portion. Some enzymes known as "exo-endonucleases", however, are not limited to either nuclease function, displaying qualities that are both endo- and exo-like. Evidence suggests that endonuclease activity experiences a lag compared to exonuclease activity.
Micrococcal nuclease is an endo-exonuclease that preferentially digests single-stranded nucleic acids. The rate of cleavage is 30 times greater at the 5' side of A or T than at G or C and results in the production of mononucleotides and oligonucleotides with terminal 3'-phosphates. The enzyme is also active against double-stranded DNA and RNA and all sequences will be ultimately cleaved.

EcoRV is a type II restriction endonuclease isolated from certain strains of Escherichia coli. It has the alternative name Eco32I.
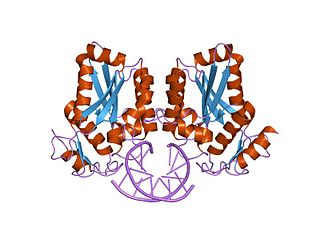
BamHI is a type II restriction endonuclease, having the capacity for recognizing short sequences of DNA and specifically cleaving them at a target site. This exhibit focuses on the structure-function relations of BamHI as described by Newman, et al. (1995). BamHI binds at the recognition sequence 5'-GGATCC-3', and cleaves these sequences just after the 5'-guanine on each strand. This cleavage results in sticky ends which are 4 bp long. In its unbound form, BamHI displays a central b sheet, which resides in between α-helices.

The homing endonucleases are a collection of endonucleases encoded either as freestanding genes within introns, as fusions with host proteins, or as self-splicing inteins. They catalyze the hydrolysis of genomic DNA within the cells that synthesize them, but do so at very few, or even singular, locations. Repair of the hydrolyzed DNA by the host cell frequently results in the gene encoding the homing endonuclease having been copied into the cleavage site, hence the term 'homing' to describe the movement of these genes. Homing endonucleases can thereby transmit their genes horizontally within a host population, increasing their allele frequency at greater than Mendelian rates.
Deoxyribonuclease IV (phage-T4-induced) is catalyzes the degradation nucleotides in DsDNA by attacking the 5'-terminal end.

BglII is a type II restriction endonuclease isolated from certain strains of Bacillus globigii.

Cystathionine beta-lyase, also commonly referred to as CBL or β-cystathionase, is an enzyme that primarily catalyzes the following α,β-elimination reaction
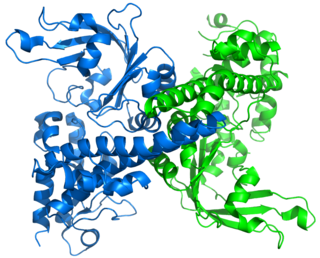
Restriction endonuclease (REase) EcoRII is an enzyme of restriction modification system (RM) naturally found in Escherichia coli, a Gram-negative bacteria. Its molecular mass is 45.2 kDa, being composed of 402 amino acids.
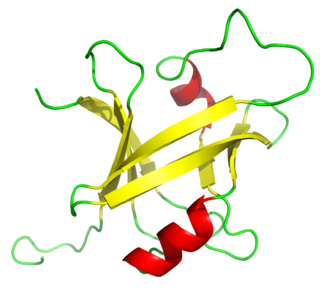
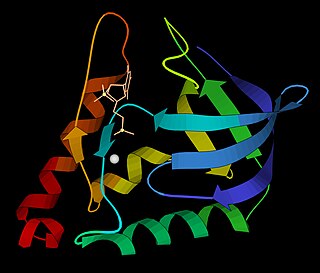
The B3 DNA binding domain (DBD) is a highly conserved domain found exclusively in transcription factors combined with other domains. It consists of 100-120 residues, includes seven beta strands and two alpha helices that form a DNA-binding pseudobarrel protein fold ; it interacts with the major groove of DNA.
PstI is a type II restriction endonuclease isolated from the Gram negative species, Providencia stuartii.

Very short patch (VSP) repair is a DNA repair system that removes GT mismatches created by the deamination of 5-methylcytosine to thymine. This system exists because the glycosylases which normally target deaminated bases cannot target thymine.

S-Adenosylmethionine synthetase, also known as methionine adenosyltransferase (MAT), is an enzyme that creates S-adenosylmethionine by reacting methionine and ATP.
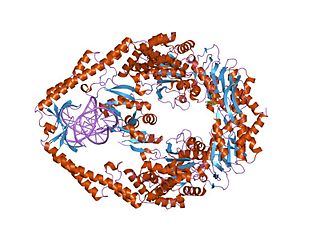
MutS is a mismatch DNA repair protein, originally described in Escherichia coli.
In molecular biology, BpuJI is a type II restriction endonuclease which recognises the asymmetric sequence 5'-CCCGT and cuts at multiple sites in the surrounding area of the target sequence. The BpuJI protein consists of two distinct modules; an N-terminal DNA recognition domain, and a C-terminal dimerisation and catalysis domain. The N-terminal domain is composed of two winged-helix subdomains and a disrupted linker subdomain. Target sequence recognition occurs through major groove contacts of amino acids in the winged-helix subdomains.

In molecular biology, the H2TH domain is a DNA-binding domain found in DNA glycosylase/AP lyase enzymes, which are involved in base excision repair of DNA damaged by oxidation or by mutagenic agents. Most damage to bases in DNA is repaired by the base excision repair pathway. These enzymes are primarily from bacteria, and have both DNA glycosylase activity EC 3.2.2.- and AP lyase activity EC 4.2.99.18. Examples include formamidopyrimidine-DNA glycosylases and endonuclease VIII (Nei).
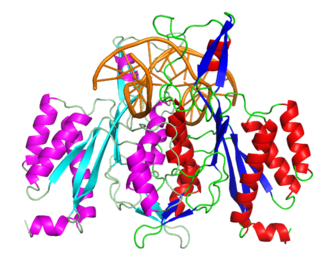
EcoRI is a restriction endonuclease enzyme isolated from species E. coli. It is a restriction enzyme that cleaves DNA double helices into fragments at specific sites, and is also a part of the restriction modification system. The Eco part of the enzyme's name originates from the species from which it was isolated - "E" denotes generic name which is "Escherichia" and "co" denotes species name, "coli" - while the R represents the particular strain, in this case RY13, and the I denotes that it was the first enzyme isolated from this strain.












