Related Research Articles
A restriction enzyme, restriction endonuclease, REase, ENase orrestrictase is an enzyme that cleaves DNA into fragments at or near specific recognition sites within molecules known as restriction sites. Restriction enzymes are one class of the broader endonuclease group of enzymes. Restriction enzymes are commonly classified into five types, which differ in their structure and whether they cut their DNA substrate at their recognition site, or if the recognition and cleavage sites are separate from one another. To cut DNA, all restriction enzymes make two incisions, once through each sugar-phosphate backbone of the DNA double helix.
Deoxyribonuclease refers to a group of glycoprotein endonucleases which are enzymes that catalyze the hydrolytic cleavage of phosphodiester linkages in the DNA backbone, thus degrading DNA. The role of the DNase enzyme in cells includes breaking down extracellular DNA (ecDNA) excreted by apoptosis, necrosis, and neutrophil extracellular traps (NET) of cells to help reduce inflammatory responses that otherwise are elicited. A wide variety of deoxyribonucleases are known and fall into one of two families, which differ in their substrate specificities, chemical mechanisms, and biological functions. Laboratory applications of DNase include purifying proteins when extracted from prokaryotic organisms. Additionally, DNase has been applied as a treatment for diseases that are caused by ecDNA in the blood plasma. Assays of DNase are emerging in the research field as well.

A nuclease is an enzyme capable of cleaving the phosphodiester bonds between nucleotides of nucleic acids. Nucleases variously effect single and double stranded breaks in their target molecules. In living organisms, they are essential machinery for many aspects of DNA repair. Defects in certain nucleases can cause genetic instability or immunodeficiency. Nucleases are also extensively used in molecular cloning.
In molecular biology, endonucleases are enzymes that cleave the phosphodiester bond within a polynucleotide chain. Some, such as deoxyribonuclease I, cut DNA relatively nonspecifically, while many, typically called restriction endonucleases or restriction enzymes, cleave only at very specific nucleotide sequences. Endonucleases differ from exonucleases, which cleave the ends of recognition sequences instead of the middle (endo) portion. Some enzymes known as "exo-endonucleases", however, are not limited to either nuclease function, displaying qualities that are both endo- and exo-like. Evidence suggests that endonuclease activity experiences a lag compared to exonuclease activity.

Exonucleases are enzymes that work by cleaving nucleotides one at a time from the end (exo) of a polynucleotide chain. A hydrolyzing reaction that breaks phosphodiester bonds at either the 3′ or the 5′ end occurs. Its close relative is the endonuclease, which cleaves phosphodiester bonds in the middle (endo) of a polynucleotide chain. Eukaryotes and prokaryotes have three types of exonucleases involved in the normal turnover of mRNA: 5′ to 3′ exonuclease (Xrn1), which is a dependent decapping protein; 3′ to 5′ exonuclease, an independent protein; and poly(A)-specific 3′ to 5′ exonuclease.
Photolyases are DNA repair enzymes that repair damage caused by exposure to ultraviolet light. These enzymes require visible light both for their own activation and for the actual DNA repair. The DNA repair mechanism involving photolyases is called photoreactivation. They mainly convert pyrimidine dimers into a normal pair of pyrimidine bases.
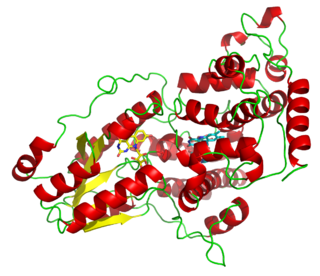
UvrABC endonuclease is a multienzyme complex in bacteria involved in DNA repair by nucleotide excision repair, and it is, therefore, sometimes called an excinuclease. This UvrABC repair process, sometimes called the short-patch process, involves the removal of twelve nucleotides where a genetic mutation has occurred followed by a DNA polymerase, replacing these aberrant nucleotides with the correct nucleotides and completing the DNA repair. The subunits for this enzyme are encoded in the uvrA, uvrB, and uvrC genes. This enzyme complex is able to repair many different types of damage, including cyclobutyl dimer formation.

Deoxyribonuclease I, is an endonuclease of the DNase family coded by the human gene DNASE1. DNase I is a nuclease that cleaves DNA preferentially at phosphodiester linkages adjacent to a pyrimidine nucleotide, yielding 5'-phosphate-terminated polynucleotides with a free hydroxyl group on position 3', on average producing tetranucleotides. It acts on single-stranded DNA, double-stranded DNA, and chromatin. In addition to its role as a waste-management endonuclease, it has been suggested to be one of the deoxyribonucleases responsible for DNA fragmentation during apoptosis.
Deoxyribonuclease V is an enzyme. This enzyme catalyses the following chemical reaction
Excision endonuclease, also known as excinuclease or UV-specific endonuclease, is a nuclease (enzyme) which excises a fragment of nucleotides during DNA repair. The excinuclease cuts out a fragment by hydrolyzing two phosphodiester bonds, one on either side of the lesion in the DNA. This process is part of "nucleotide excision repair", a mechanism that can fix specific types of damage to the DNA in the G1 phase of the eukaryotic cell cycle. Such damage may include thymine dimers created by UV rays as well as the bulky distortions in DNA caused by oxidized benzopyrenes from sources such as cigarette smoke.
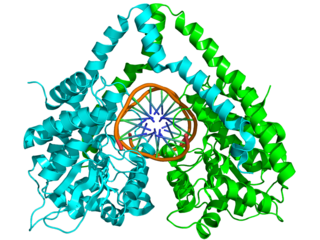
HindIII (pronounced "Hin D Three") is a type II site-specific deoxyribonuclease restriction enzyme isolated from Haemophilus influenzae that cleaves the DNA palindromic sequence AAGCTT in the presence of the cofactor Mg2+ via hydrolysis.
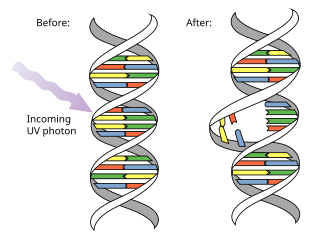
Pyrimidine dimers are molecular lesions formed from thymine or cytosine bases in DNA via photochemical reactions, commonly associated with direct DNA damage. Ultraviolet light induces the formation of covalent linkages between consecutive bases along the nucleotide chain in the vicinity of their carbon–carbon double bonds. The photo-coupled dimers are fluorescent. The dimerization reaction can also occur among pyrimidine bases in dsRNA —uracil or cytosine. Two common UV products are cyclobutane pyrimidine dimers (CPDs) and 6–4 photoproducts. These premutagenic lesions alter the structure of the DNA helix and cause non-canonical base pairing. Specifically, adjacent thymines or cytosines in DNA will form a cyclobutane ring when joined together and cause a distortion in the DNA. This distortion prevents replication or transcription machinery beyond the site of the dimerization. Up to 50–100 such reactions per second might occur in a skin cell during exposure to sunlight, but are usually corrected within seconds by photolyase reactivation or nucleotide excision repair. In humans, the most common form of DNA repair is nucleotide excision repair (NER). In contrast, organisms such as bacteria can counterintuitively harvest energy from the sun to fix DNA damage from pyrimidine dimers via photolyase activity. If these lesions are not fixed, polymerase machinery may misread or add in the incorrect nucleotide to the strand. If the damage to the DNA is overwhelming, mutations can arise within the genome of an organism and may lead to the production of cancer cells. Uncorrected lesions can inhibit polymerases, cause misreading during transcription or replication, or lead to arrest of replication. It causes sunburn and it triggers the production of melanin. Pyrimidine dimers are the primary cause of melanomas in humans.
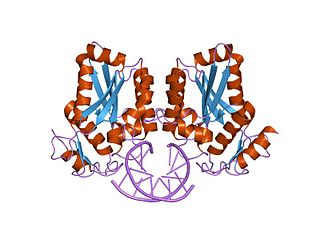
BamHI is a type II restriction endonuclease, having the capacity for recognizing short sequences of DNA and specifically cleaving them at a target site. This exhibit focuses on the structure-function relations of BamHI as described by Newman, et al. (1995). BamHI binds at the recognition sequence 5'-GGATCC-3', and cleaves these sequences just after the 5'-guanine on each strand. This cleavage results in sticky ends which are 4 bp long. In its unbound form, BamHI displays a central b sheet, which resides in between α-helices.

Nuclease S1 is an endonuclease enzyme that splits single-stranded DNA (ssDNA) and RNA into oligo- or mononucleotides. This enzyme catalyses the following chemical reaction
Deoxyribonuclease IV (phage-T4-induced) is catalyzes the degradation nucleotides in DsDNA by attacking the 5'-terminal end.
The enzyme DNA-(apurinic or apyrimidinic site) lyase, also referred to as DNA-(apurinic or apyrimidinic site) 5'-phosphomonoester-lyase or DNA AP lyase catalyzes the cleavage of the C-O-P bond 3' from the apurinic or apyrimidinic site in DNA via β-elimination reaction, leaving a 3'-terminal unsaturated sugar and a product with a terminal 5'-phosphate. In the 1970s, this class of enzyme was found to repair at apurinic or apyrimidinic DNA sites in E. coli and in mammalian cells. The major active enzyme of this class in bacteria, and specifically, E. coli is endonuclease type III. This enzyme is part of a family of lyases that cleave carbon-oxygen bonds.
The term proofreading is used in genetics to refer to the error-correcting processes, first proposed by John Hopfield and Jacques Ninio, involved in DNA replication, immune system specificity, enzyme-substrate recognition among many other processes that require enhanced specificity. The proofreading mechanisms of Hopfield and Ninio are non-equilibrium active processes that consume ATP to enhance specificity of various biochemical reactions.
Crossover junction endodeoxyribonuclease, also known as Holliday junction resolvase, Holliday junction endonuclease, Holliday junction-cleaving endonuclease, Holliday junction-resolving endoribonuclease, crossover junction endoribonuclease, and cruciform-cutting endonuclease, is an enzyme involved in DNA repair and homologous recombination. Specifically, it performs endonucleolytic cleavage that results in single-stranded crossover between two homologous DNA molecules at the Holliday junction to produce recombinant DNA products for chromosomal segregation. This process is known as Holliday junction resolution.
Deoxyribodipyrimidine endonucleosidase is an enzyme with systematic name deoxy-D-ribocyclobutadipyrimidine polynucleotidodeoxyribohydrolase. This enzyme catalyses the following chemical reaction
References
- ↑ Braun AG, Radman M, Grossman L (September 1976). "Enzymatic repair of DNA: SITES OF HYDROLYSIS BY THE Escherichia coli endonuclease specific for pyrimidine dimers (correndonuclease II)". Biochemistry. 15 (18): 4116–20. doi:10.1021/bi00663a031. PMID 786366.
- ↑ Riazuddin S, Grossman L (September 1977). "Micrococcus luteus correndonucleases. II. Mechanism of action of two endonucleases specific for DNA containing pyrimidine dimers". The Journal of Biological Chemistry. 252 (18): 6287–93. doi: 10.1016/S0021-9258(17)39953-2 . PMID 330526.