Related Research Articles
A restriction enzyme, restriction endonuclease, REase, ENase orrestrictase is an enzyme that cleaves DNA into fragments at or near specific recognition sites within molecules known as restriction sites. Restriction enzymes are one class of the broader endonuclease group of enzymes. Restriction enzymes are commonly classified into five types, which differ in their structure and whether they cut their DNA substrate at their recognition site, or if the recognition and cleavage sites are separate from one another. To cut DNA, all restriction enzymes make two incisions, once through each sugar-phosphate backbone of the DNA double helix.

Ribonuclease is a type of nuclease that catalyzes the degradation of RNA into smaller components. Ribonucleases can be divided into endoribonucleases and exoribonucleases, and comprise several sub-classes within the EC 2.7 and 3.1 classes of enzymes.

Ribonuclease H is a family of non-sequence-specific endonuclease enzymes that catalyze the cleavage of RNA in an RNA/DNA substrate via a hydrolytic mechanism. Members of the RNase H family can be found in nearly all organisms, from bacteria to archaea to eukaryotes.

Transcriptional modification or co-transcriptional modification is a set of biological processes common to most eukaryotic cells by which an RNA primary transcript is chemically altered following transcription from a gene to produce a mature, functional RNA molecule that can then leave the nucleus and perform any of a variety of different functions in the cell. There are many types of post-transcriptional modifications achieved through a diverse class of molecular mechanisms.
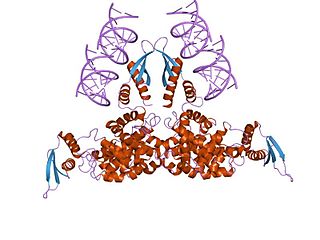
Ribonuclease III (RNase III or RNase C)(BRENDA 3.1.26.3) is a type of ribonuclease that recognizes dsRNA and cleaves it at specific targeted locations to transform them into mature RNAs. These enzymes are a group of endoribonucleases that are characterized by their ribonuclease domain, which is labelled the RNase III domain. They are ubiquitous compounds in the cell and play a major role in pathways such as RNA precursor synthesis, RNA Silencing, and the pnp autoregulatory mechanism.

Inositol oxygenase, also commonly referred to as myo-inositol oxygenase (MIOX), is a non-heme di-iron enzyme that oxidizes myo-inositol to glucuronic acid. The enzyme employs a unique four-electron transfer at its Fe(II)/Fe(III) coordination sites and the reaction proceeds through the direct binding of myo-inositol followed by attack of the iron center by diatomic oxygen. This enzyme is part of the only known pathway for the catabolism of inositol in humans and is expressed primarily in the kidneys. Recent medical research regarding MIOX has focused on understanding its role in metabolic and kidney diseases such as diabetes, obesity and acute kidney injury. Industrially-focused engineering efforts are centered on improving MIOX activity in order to produce glucaric acid in heterologous hosts.

Transaldolase is an enzyme of the non-oxidative phase of the pentose phosphate pathway. In humans, transaldolase is encoded by the TALDO1 gene.

An exoribonuclease is an exonuclease ribonuclease, which are enzymes that degrade RNA by removing terminal nucleotides from either the 5' end or the 3' end of the RNA molecule. Enzymes that remove nucleotides from the 5' end are called 5'-3' exoribonucleases, and enzymes that remove nucleotides from the 3' end are called 3'-5' exoribonucleases.
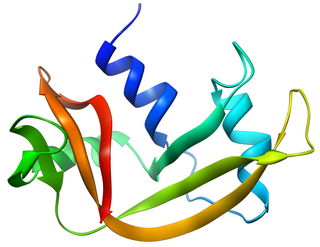
Pancreatic ribonuclease family is a superfamily of pyrimidine-specific endonucleases found in high quantity in the pancreas of certain mammals and of some reptiles.
Exoribonuclease II is an enzyme. This enzyme catalyses the following chemical reaction
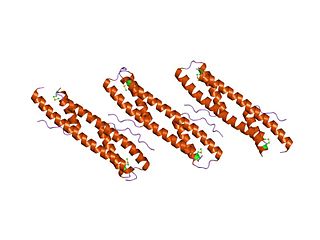
The enzyme exodeoxyribonuclease VII is a bacterial exonuclease enzyme. It is composed of two nonidentical subunits; one large subunit and 4 small ones. that catalyses exonucleolytic cleavage in either 5′- to 3′- or 3′- to 5′-direction to yield nucleoside 5′-phosphates. The large subunit also contains an N-terminal OB-fold domain that binds to nucleic acids.
Ribonuclease T2 is an enzyme. It is a type of endoribonuclease. This enzyme catalyses the following chemical reaction
Ribonuclease IV is an enzyme. This enzyme catalyses the following chemical reaction
Ribonuclease is an enzyme. This enzyme catalyses the following chemical reaction
Ribonuclease IX is an enzyme. This enzyme catalyses the following chemical reaction
Ribonuclease E is a bacterial ribonuclease that participates in the processing of ribosomal RNA and the chemical degradation of bulk cellular RNA.
Ribonuclease U2 is an enzyme. This enzyme catalyses the following chemical reaction
Ribonuclease F is an enzyme. This enzyme catalyses the following chemical reaction

RNA hydrolysis is a reaction in which a phosphodiester bond in the sugar-phosphate backbone of RNA is broken, cleaving the RNA molecule. RNA is susceptible to this base-catalyzed hydrolysis because the ribose sugar in RNA has a hydroxyl group at the 2’ position. This feature makes RNA chemically unstable compared to DNA, which does not have this 2’ -OH group and thus is not susceptible to base-catalyzed hydrolysis.

Ribonuclease T is a ribonuclease enzyme involved in the maturation of transfer RNA and ribosomal RNA in bacteria, as well as in DNA repair pathways. It is a member of the DnaQ family of exonucleases and non-processively acts on the 3' end of single-stranded nucleic acids. RNase T is capable of cleaving both DNA and RNA, with extreme sequence specificity discriminating against cytosine at the 3' end of the substrate.
References
- ↑ Levy CC, Goldman P (June 1970). "Residue specificity of a ribonuclease which hydrolyzes polycytidylic acid". The Journal of Biological Chemistry. 245 (12): 3257–62. doi: 10.1016/S0021-9258(18)63048-0 . PMID 5432809.
- ↑ Marotta CA, Levy CC, Weissman SM, Varricchio F (July 1973). "Preferred sites of digestion of a ribonuclease from Enterobacter sp. in the sequence analysis of Bacillus stearothermophilus 5S ribonucleic acid". Biochemistry. 12 (15): 2901–4. doi:10.1021/bi00739a020. PMID 4719125.