Related Research Articles

A DNA polymerase is a member of a family of enzymes that catalyze the synthesis of DNA molecules from nucleoside triphosphates, the molecular precursors of DNA. These enzymes are essential for DNA replication and usually work in groups to create two identical DNA duplexes from a single original DNA duplex. During this process, DNA polymerase "reads" the existing DNA strands to create two new strands that match the existing ones. These enzymes catalyze the chemical reaction

DNA polymerase I is an enzyme that participates in the process of prokaryotic DNA replication. Discovered by Arthur Kornberg in 1956, it was the first known DNA polymerase. It was initially characterized in E. coli and is ubiquitous in prokaryotes. In E. coli and many other bacteria, the gene that encodes Pol I is known as polA. The E. coli Pol I enzyme is composed of 928 amino acids, and is an example of a processive enzyme — it can sequentially catalyze multiple polymerisation steps without releasing the single-stranded template. The physiological function of Pol I is mainly to support repair of damaged DNA, but it also contributes to connecting Okazaki fragments by deleting RNA primers and replacing the ribonucleotides with DNA.
In molecular biology, endonucleases are enzymes that cleave the phosphodiester bond within a polynucleotide chain. Some, such as deoxyribonuclease I, cut DNA relatively nonspecifically, while many, typically called restriction endonucleases or restriction enzymes, cleave only at very specific nucleotide sequences. Endonucleases differ from exonucleases, which cleave the ends of recognition sequences instead of the middle (endo) portion. Some enzymes known as "exo-endonucleases", however, are not limited to either nuclease function, displaying qualities that are both endo- and exo-like. Evidence suggests that endonuclease activity experiences a lag compared to exonuclease activity.

Exonucleases are enzymes that work by cleaving nucleotides one at a time from the end (exo) of a polynucleotide chain. A hydrolyzing reaction that breaks phosphodiester bonds at either the 3′ or the 5′ end occurs. Its close relative is the endonuclease, which cleaves phosphodiester bonds in the middle (endo) of a polynucleotide chain. Eukaryotes and prokaryotes have three types of exonucleases involved in the normal turnover of mRNA: 5′ to 3′ exonuclease (Xrn1), which is a dependent decapping protein; 3′ to 5′ exonuclease, an independent protein; and poly(A)-specific 3′ to 5′ exonuclease.

DNA polymerase II is a prokaryotic DNA-dependent DNA polymerase encoded by the PolB gene.

Exonuclease III (ExoIII) is an enzyme that belongs to the exonuclease family. ExoIII catalyzes the stepwise removal of mononucleotides from 3´-hydroxyl termini of double-stranded DNA. A limited number of nucleotides are removed during each binding event, resulting in coordinated progressive deletions within the population of DNA molecules.
Exodeoxyribonuclease I is an enzyme that catalyses the following chemical reaction:
Purple acid phosphatases (PAPs) (EC 3.1.3.2) are metalloenzymes that hydrolyse phosphate esters and anhydrides under acidic condition. In their oxidised form, PAPs in solution are purple in colour. This is due to the presence of a dinuclear iron centre, to which a tyrosine residue is connected via a charge transfer. This metallic centre is composed of Fe3+ and M, where M is Fe3+, Zn2+, Mg2+ or Mn2+. The conserved Fe3+ is stabilised in the ferric form, whereas M may undergo reduction. Upon treatment with mild reductants, PAPs are converted to their enzymatically active, pink form. Treatment with strong reducing agents dissociates the metallic ions, and renders the enzyme colourless and inactive.

Polynucleotide Phosphorylase (PNPase) is a bifunctional enzyme with a phosphorolytic 3' to 5' exoribonuclease activity and a 3'-terminal oligonucleotide polymerase activity. That is, it dismantles the RNA chain starting at the 3' end and working toward the 5' end. It also synthesizes long, highly heteropolymeric tails in vivo. It accounts for all of the observed residual polyadenylation in strains of Escherichia coli missing the normal polyadenylation enzyme. Discovered by Marianne Grunberg-Manago working in Severo Ochoa's lab in 1955, the RNA-polymerization activity of PNPase was initially believed to be responsible for DNA-dependent synthesis of messenger RNA, a notion that was disproven by the late 1950s.
Deoxyribonuclease IV (phage-T4-induced) is catalyzes the degradation nucleotides in DsDNA by attacking the 5'-terminal end.
In enzymology, a [acyl-carrier-protein] S-malonyltransferase is an enzyme that catalyzes the chemical reaction

In enzymology, a cystathionine gamma-synthase is an enzyme that catalyzes the formation of cystathionine from cysteine and an activated derivative of homoserine, e.g.:

T7 DNA polymerase is an enzyme used during the DNA replication of the T7 bacteriophage. During this process, the DNA polymerase “reads” existing DNA strands and creates two new strands that match the existing ones. The T7 DNA polymerase requires a host factor, E. coli thioredoxin, in order to carry out its function. This helps stabilize the binding of the necessary protein to the primer-template to improve processivity by more than 100-fold, which is a feature unique to this enzyme. It is a member of the Family A DNA polymerases, which include E. coli DNA polymerase I and Taq DNA polymerase.
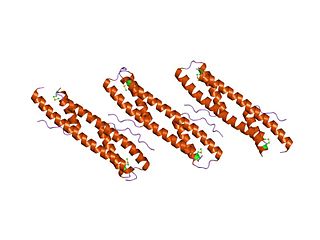
The enzyme exodeoxyribonuclease VII is a bacterial exonuclease enzyme. It is composed of two nonidentical subunits; one large subunit and 4 small ones. that catalyses exonucleolytic cleavage in either 5′- to 3′- or 3′- to 5′-direction to yield nucleoside 5′-phosphates. The large subunit also contains an N-terminal OB-fold domain that binds to nucleic acids.
Serratia marcescens nuclease is an enzyme. This enzyme catalyses the following chemical reaction
Exodeoxyribonuclease (lambda-induced) is an exonuclease. This enzyme catalyses the following chemical reaction
Pitrilysin is an enzyme. This enzyme catalyses the following chemical reaction:

Ribonuclease T is a ribonuclease enzyme involved in the maturation of transfer RNA and ribosomal RNA in bacteria, as well as in DNA repair pathways. It is a member of the DnaQ family of exonucleases and non-processively acts on the 3' end of single-stranded nucleic acids. RNase T is capable of cleaving both DNA and RNA, with extreme sequence specificity discriminating against cytosine at the 3' end of the substrate.
Sylvy Kornberg née Sylvia Ruth Levy (1917–1986) was an American biochemist who carried out research on DNA replication and polyphosphate synthesis. She discovered and characterized polyphosphate kinase (PPK), an enzyme that helps build long chains of phosphate groups called polyphosphate (PolyP) that play a variety of metabolic and regulatory functions. She worked closely with her husband and research partner, Arthur Kornberg, contributing greatly to the characterization of DNA polymerization that earned him the 1959 Nobel Prize in Physiology or Medicine.
Charles Clifton Richardson is an American biochemist and professor at Harvard University. Richardson received his undergraduate education at Duke University, where he majored in medicine. He received his M.D. at Duke Medical School in 1960. Richardson works as a professor at Harvard Medical School, and he served as editor/associate editor of the Annual Review of Biochemistry from 1972 to 2003. Richardson received the American Chemical Society Award in Biological Chemistry in 1968, as well as numerous other accolades.
References
- ↑ Lindahl T, Gally JA, Edelman GM (September 1969). "Properties of deoxyribonuclease 3 from mammalian tissues". The Journal of Biological Chemistry. 244 (18): 5014–9. doi: 10.1016/S0021-9258(18)94303-6 . PMID 5824576.
- ↑ Richardson CC, Kornberg A (January 1964). "A deoxyribonucleic acid phosphatase-exonuclease from Escherichia coli. I. Purification of the enzyme and characterization of the phosphatase activity". The Journal of Biological Chemistry. 239: 242–50. doi: 10.1016/S0021-9258(18)51774-9 . PMID 14114850.
- ↑ Richardson CC, Lehman IR, Kornberg A (January 1964). "A deoxyribonucleic acid phosphatase-exonuclease from Escherichia coli. II. Characterization of the exonuclease activity". The Journal of Biological Chemistry. 239: 251–8. doi: 10.1016/S0021-9258(18)51775-0 . PMID 14114851.