Related Research Articles

Proteoglycans are proteins that are heavily glycosylated. The basic proteoglycan unit consists of a "core protein" with one or more covalently attached glycosaminoglycan (GAG) chain(s). The point of attachment is a serine (Ser) residue to which the glycosaminoglycan is joined through a tetrasaccharide bridge. The Ser residue is generally in the sequence -Ser-Gly-X-Gly-, although not every protein with this sequence has an attached glycosaminoglycan. The chains are long, linear carbohydrate polymers that are negatively charged under physiological conditions due to the occurrence of sulfate and uronic acid groups. Proteoglycans occur in connective tissue.
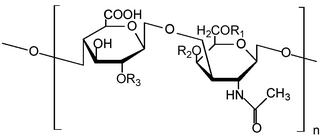
Glycosaminoglycans (GAGs) or mucopolysaccharides are long, linear polysaccharides consisting of repeating disaccharide units. The repeating two-sugar unit consists of a uronic sugar and an amino sugar, except in the case of the sulfated glycosaminoglycan keratan, where, in place of the uronic sugar there is a galactose unit. GAGs are found in vertebrates, invertebrates and bacteria. Because GAGs are highly polar molecules and attract water; the body uses them as lubricants or shock absorbers.

Heparan sulfate (HS) is a linear polysaccharide found in all animal tissues. It occurs as a proteoglycan in which two or three HS chains are attached in close proximity to cell surface or extracellular matrix proteins. In this form, HS binds to a variety of protein ligands, including Wnt, and regulates a wide range of biological activities, including developmental processes, angiogenesis, blood coagulation, abolishing detachment activity by GrB, and tumour metastasis. HS has also been shown to serve as cellular receptor for a number of viruses, including the respiratory syncytial virus. One study suggests that cellular heparan sulfate has a role in SARS-CoV-2 Infection, particularly when the virus attaches with ACE2.
In biochemistry, sulfatases EC 3.1.6.- are a class of enzymes of the esterase class that catalyze the hydrolysis of sulfate esters into an alcohol and a bisulfate:

l-Iduronic acid is the major uronic acid component of the glycosaminoglycans (GAGs) dermatan sulfate, and heparin. It is also present in heparan sulfate, although here in a minor amount relative to its carbon-5 epimer glucuronic acid.

Arylsulfatase B is an enzyme associated with mucopolysaccharidosis VI.

N-acetylgalactosamine-6-sulfatase is an enzyme that, in humans, is encoded by the GALNS gene.

N-acetylglucosamine-6-sulfatase (EC 3.1.6.14, glucosamine (N-acetyl)-6-sulfatase, systematic name N-acetyl-D-glucosamine-6-sulfate 6-sulfohydrolase) is an enzyme that in humans is encoded by the GNS gene. It is deficient in Sanfilippo Syndrome type IIId. It catalyses the hydrolysis of the 6-sulfate groups of the N-acetyl-D-glucosamine 6-sulfate units of heparan sulfate and keratan sulfate
Heparosan-N-sulfate-glucuronate 5-epimerase is an enzyme with systematic name poly( -beta-D-glucuronosyl- -N-sulfo-alpha-D-glucosaminyl) glucurono-5-epimerase. This enzyme catalyses the following chemical reaction

In enzymology, a N-sulfoglucosamine sulfohydrolase (EC 3.10.1.1), otherwise known as SGSH, is an enzyme that catalyzes the chemical reaction
The enzyme heparin lyase catalyzes the following process:
The enzyme disulfoglucosamine-6-sulfatase (EC 3.1.6.1) catalyzes the reaction
The enzyme N-acetylgalactosamine-6-sulfatase catalyzes the chemical reaction of cleaving off the 6-sulfate groups of the N-acetyl-D-galactosamine 6-sulfate units of the macromolecule chondroitin sulfate and, similarly, of the D-galactose 6-sulfate units of the macromolecule keratan sulfate.
The enzyme N-sulfoglucosamine-3-sulfatase catalyzes cleaving off the 3-sulfate groups of the N-sulfo-D-glucosamine 3-O-sulfate units of heparin.
In enzymology, a galactose-6-sulfurylase is an enzyme that catalyzes the chemical reaction

In enzymology, a galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase is an enzyme that catalyzes the chemical reaction

Sulfatase-modifying factor 1 is an enzyme that in humans is encoded by the SUMF1 gene.

Galactose-3-O-sulfotransferase 4 is an enzyme that in humans is encoded by the GAL3ST4 gene.

Formylglycine-generating enzyme (FGE), located at 3p26.1 in humans, is the name for an enzyme present in the endoplasmic reticulum that catalyzes the conversion of cysteine to formylglycine (fGly). There are two main classes of FGE, aerobic and anaerobic. FGE activates sulfatases, which are essential for the degradation of sulfate esters. The catalytic activity of sulfatases is dependent upon a formylglycine residue in the active site.
Unsaturated chondroitin disaccharide hydrolase (EC 3.2.1.180, UGL, unsaturated glucuronyl hydrolase) is an enzyme with systematic name beta-D-4-deoxy-Delta4-GlcAp-(1->3)-beta-D-GalNAc6S hydrolase. This enzyme catalyses the following chemical reaction
References
- Shaklee PN, Glaser JH, Conrad HE (1985). "A sulfatase specific for glucuronic acid 2-sulfate residues in glycosaminoglycans". J. Biol. Chem. 260 (16): 9146–9. PMID 4019466.