
Photosynthesis is a biological process used by many cellular organisms to convert light energy into chemical energy, which is stored in organic compounds that can later be metabolized through cellular respiration to fuel the organism's activities. The term usually refers to oxygenic photosynthesis, where oxygen is produced as a byproduct and some of the chemical energy produced is stored in carbohydrate molecules such as sugars, starch, glycogen and cellulose, which are synthesized from endergonic reaction of carbon dioxide with water. Most plants, algae and cyanobacteria perform photosynthesis; such organisms are called photoautotrophs. Photosynthesis is largely responsible for producing and maintaining the oxygen content of the Earth's atmosphere, and supplies most of the biological energy necessary for complex life on Earth.

The green sulfur bacteria are a phylum, Chlorobiota, of obligately anaerobic photoautotrophic bacteria that metabolize sulfur.

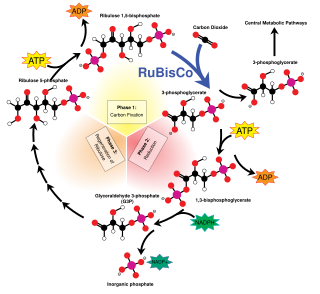
Ribulose-1,5-bisphosphate carboxylase/oxygenase, commonly known by the abbreviations RuBisCo, rubisco, RuBPCase, or RuBPco, is an enzyme involved in light-independent part of photosynthesis, including the carbon fixation by which atmospheric carbon dioxide is converted by plants and other photosynthetic organisms to energy-rich molecules such as glucose. It emerged approximately four billion years ago in primordial metabolism prior to the presence of oxygen on earth. It is probably the most abundant enzyme on Earth. In chemical terms, it catalyzes the carboxylation of ribulose-1,5-bisphosphate.

Photorespiration (also known as the oxidative photosynthetic carbon cycle or C2 cycle) refers to a process in plant metabolism where the enzyme RuBisCO oxygenates RuBP, wasting some of the energy produced by photosynthesis. The desired reaction is the addition of carbon dioxide to RuBP (carboxylation), a key step in the Calvin–Benson cycle, but approximately 25% of reactions by RuBisCO instead add oxygen to RuBP (oxygenation), creating a product that cannot be used within the Calvin–Benson cycle. This process lowers the efficiency of photosynthesis, potentially lowering photosynthetic output by 25% in C3 plants. Photorespiration involves a complex network of enzyme reactions that exchange metabolites between chloroplasts, leaf peroxisomes and mitochondria.

Ribulose 1,5-bisphosphate (RuBP) is an organic substance that is involved in photosynthesis, notably as the principal CO2 acceptor in plants. It is a colourless anion, a double phosphate ester of the ketopentose called ribulose. Salts of RuBP can be isolated, but its crucial biological function happens in solution. RuBP occurs not only in plants but in all domains of life, including Archaea, Bacteria, and Eukarya.
C3 carbon fixation is the most common of three metabolic pathways for carbon fixation in photosynthesis, the other two being C4 and CAM. This process converts carbon dioxide and ribulose bisphosphate (RuBP, a 5-carbon sugar) into two molecules of 3-phosphoglycerate through the following reaction:
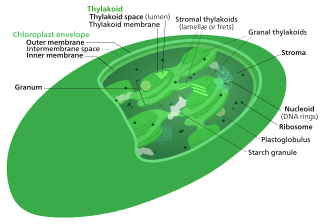
The Calvin cycle,light-independent reactions, bio synthetic phase,dark reactions, or photosynthetic carbon reduction (PCR) cycle of photosynthesis is a series of chemical reactions that convert carbon dioxide and hydrogen-carrier compounds into glucose. The Calvin cycle is present in all photosynthetic eukaryotes and also many photosynthetic bacteria. In plants, these reactions occur in the stroma, the fluid-filled region of a chloroplast outside the thylakoid membranes. These reactions take the products of light-dependent reactions and perform further chemical processes on them. The Calvin cycle uses the chemical energy of ATP and reducing power of NADPH from the light dependent reactions to produce sugars for the plant to use. These substrates are used in a series of reduction-oxidation reactions to produce sugars in a step-wise process; there is no direct reaction that converts several molecules of CO2 to a sugar. There are three phases to the light-independent reactions, collectively called the Calvin cycle: carboxylation, reduction reactions, and ribulose 1,5-bisphosphate (RuBP) regeneration.
Dihydroxyacetone phosphate (DHAP, also glycerone phosphate in older texts) is the anion with the formula HOCH2C(O)CH2OPO32-. This anion is involved in many metabolic pathways, including the Calvin cycle in plants and glycolysis. It is the phosphate ester of dihydroxyacetone.

DCMU is an algicide and herbicide of the arylurea class that inhibits photosynthesis. It was introduced by Bayer in 1954 under the trade name of Diuron.
Phosphoenolpyruvate carboxylase (also known as PEP carboxylase, PEPCase, or PEPC; EC 4.1.1.31, PDB ID: 3ZGE) is an enzyme in the family of carboxy-lyases found in plants and some bacteria that catalyzes the addition of bicarbonate (HCO3−) to phosphoenolpyruvate (PEP) to form the four-carbon compound oxaloacetate and inorganic phosphate:

Sugar phosphates are often used in biological systems to store or transfer energy. They also form the backbone for DNA and RNA. Sugar phosphate backbone geometry is altered in the vicinity of the modified nucleotides.
Peroxiredoxins are a ubiquitous family of antioxidant enzymes that also control cytokine-induced peroxide levels and thereby mediate signal transduction in mammalian cells. The family members in humans are PRDX1, PRDX2, PRDX3, PRDX4, PRDX5, and PRDX6. The physiological importance of peroxiredoxins is indicated by their relative abundance. Their function is the reduction of peroxides, specifically hydrogen peroxide, alkyl hydroperoxides, and peroxynitrite.

Fructose 2,6-bisphosphate, abbreviated Fru-2,6-P2, is a metabolite that allosterically affects the activity of the enzymes phosphofructokinase 1 (PFK-1) and fructose 1,6-bisphosphatase (FBPase-1) to regulate glycolysis and gluconeogenesis. Fru-2,6-P2 itself is synthesized and broken down by the bifunctional enzyme phosphofructokinase 2/fructose-2,6-bisphosphatase (PFK-2/FBPase-2).

Fructose-bisphosphate aldolase, often just aldolase, is an enzyme catalyzing a reversible reaction that splits the aldol, fructose 1,6-bisphosphate, into the triose phosphates dihydroxyacetone phosphate (DHAP) and glyceraldehyde 3-phosphate (G3P). Aldolase can also produce DHAP from other (3S,4R)-ketose 1-phosphates such as fructose 1-phosphate and sedoheptulose 1,7-bisphosphate. Gluconeogenesis and the Calvin cycle, which are anabolic pathways, use the reverse reaction. Glycolysis, a catabolic pathway, uses the forward reaction. Aldolase is divided into two classes by mechanism.

Malate dehydrogenase (oxaloacetate-decarboxylating) (NADP+) (EC 1.1.1.40) or NADP-malic enzyme (NADP-ME) is an enzyme that catalyzes the chemical reaction in the presence of a bivalent metal ion:
In enzymology, a ferredoxin-NADP+ reductase (EC 1.18.1.2) abbreviated FNR, is an enzyme that catalyzes the chemical reaction
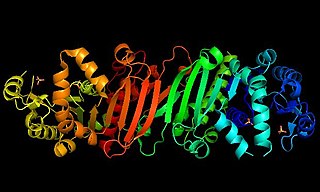
Phosphoribulokinase (PRK) (EC 2.7.1.19) is an essential photosynthetic enzyme that catalyzes the ATP-dependent phosphorylation of ribulose 5-phosphate (RuP) into ribulose 1,5-bisphosphate (RuBP), both intermediates in the Calvin Cycle. Its main function is to regenerate RuBP, which is the initial substrate and CO2-acceptor molecule of the Calvin Cycle. PRK belongs to the family of transferase enzymes, specifically those transferring phosphorus-containing groups (phosphotransferases) to an alcohol group acceptor. Along with ribulose 1,5-bisphosphate carboxylase/oxygenase (RuBisCo), phosphoribulokinase is unique to the Calvin Cycle. Therefore, PRK activity often determines the metabolic rate in organisms for which carbon fixation is key to survival. Much initial work on PRK was done with spinach leaf extracts in the 1950s; subsequent studies of PRK in other photosynthetic prokaryotic and eukaryotic organisms have followed. The possibility that PRK might exist was first recognized by Weissbach et al. in 1954; for example, the group noted that carbon dioxide fixation in crude spinach extracts was enhanced by the addition of ATP. The first purification of PRK was conducted by Hurwitz and colleagues in 1956.
ATP + Mg2+ - D-ribulose 5-phosphate ADP + D-ribulose 1,5-bisphosphate
Non-photochemical quenching (NPQ) is a mechanism employed by plants and algae to protect themselves from the adverse effects of high light intensity. It involves the quenching of singlet excited state chlorophylls (Chl) via enhanced internal conversion to the ground state, thus harmlessly dissipating excess excitation energy as heat through molecular vibrations. NPQ occurs in almost all photosynthetic eukaryotes, and helps to regulate and protect photosynthesis in environments where light energy absorption exceeds the capacity for light utilization in photosynthesis.

Ferredoxin-thioredoxin reductase EC 1.8.7.2, systematic name ferredoxin:thioredoxin disulfide oxidoreductase, is a [4Fe-4S] protein that plays an important role in the ferredoxin/thioredoxin regulatory chain. It catalyzes the following reaction:

2-Phosphoglycolate (chemical formula C2H2O6P3-; also known as phosphoglycolate, 2-PG, or PG) is a natural metabolic product of the oxygenase reaction mediated by the enzyme ribulose 1,5-bisphosphate carboxylase (RuBisCo).




















