
Ribonuclease is a type of nuclease that catalyzes the degradation of RNA into smaller components. Ribonucleases can be divided into endoribonucleases and exoribonucleases, and comprise several sub-classes within the EC 2.7 and 3.1 classes of enzymes.

In biochemistry, a nuclease is an enzyme capable of cleaving the phosphodiester bonds between nucleotides of nucleic acids. Nucleases variously effect single and double stranded breaks in their target molecules. In living organisms, they are essential machinery for many aspects of DNA repair. Defects in certain nucleases can cause genetic instability or immunodeficiency. Nucleases are also extensively used in molecular cloning.

The Klenow fragment is a large protein fragment produced when DNA polymerase I from E. coli is enzymatically cleaved by the protease subtilisin. First reported in 1970, it retains the 5' → 3' polymerase activity and the 3’ → 5’ exonuclease activity for removal of precoding nucleotides and proofreading, but loses its 5' → 3' exonuclease activity.
In molecular biology, endonucleases are enzymes that cleave the phosphodiester bond within a polynucleotide chain. Some, such as deoxyribonuclease I, cut DNA relatively nonspecifically, while many, typically called restriction endonucleases or restriction enzymes, cleave only at very specific nucleotide sequences. Endonucleases differ from exonucleases, which cleave the ends of recognition sequences instead of the middle (endo) portion. Some enzymes known as "exo-endonucleases", however, are not limited to either nuclease function, displaying qualities that are both endo- and exo-like. Evidence suggests that endonuclease activity experiences a lag compared to exonuclease activity.

Exonucleases are enzymes that work by cleaving nucleotides one at a time from the end (exo) of a polynucleotide chain. A hydrolyzing reaction that breaks phosphodiester bonds at either the 3′ or the 5′ end occurs. Its close relative is the endonuclease, which cleaves phosphodiester bonds in the middle (endo) of a polynucleotide chain. Eukaryotes and prokaryotes have three types of exonucleases involved in the normal turnover of mRNA: 5′ to 3′ exonuclease (Xrn1), which is a dependent decapping protein; 3′ to 5′ exonuclease, an independent protein; and poly(A)-specific 3′ to 5′ exonuclease.
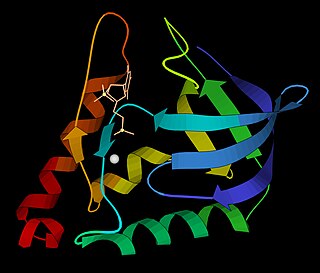
Micrococcal nuclease is an endo-exonuclease that preferentially digests single-stranded nucleic acids. The rate of cleavage is 30 times greater at the 5' side of A or T than at G or C and results in the production of mononucleotides and oligonucleotides with terminal 3'-phosphates. The enzyme is also active against double-stranded DNA and RNA and all sequences will be ultimately cleaved.

Apurinic/apyrimidinic (AP) endonuclease is an enzyme that is involved in the DNA base excision repair pathway (BER). Its main role in the repair of damaged or mismatched nucleotides in DNA is to create a nick in the phosphodiester backbone of the AP site created when DNA glycosylase removes the damaged base.

Exonuclease III (ExoIII) is an enzyme that belongs to the exonuclease family. ExoIII catalyzes the stepwise removal of mononucleotides from 3´-hydroxyl termini of double-stranded DNA. A limited number of nucleotides are removed during each binding event, resulting in coordinated progressive deletions within the population of DNA molecules.
In biochemistry, an esterase is a class of enzyme that splits esters into an acid and an alcohol in a chemical reaction with water called hydrolysis.
Mung bean nuclease is a nuclease derived from sprouts of the mung bean that removes nucleotides in a step-wise manner from single-stranded DNA molecules (ssDNA) and is used in biotechnological applications to remove such ssDNA from a mixture also containing double-stranded DNA (dsDNA). This enzyme is useful for transcript mapping, removal of single-stranded regions in DNA hybrids or single-stranded overhangs produced by restriction enzymes, etc. It has an activity similar to Nuclease S1, but it has higher specificity for single-stranded molecules.
Deoxyribonuclease II is an endonuclease that hydrolyzes phosphodiester linkages of deoxyribonucleotide in native and denatured DNA, yielding products with 3'-phosphates and 5'-hydroxyl ends, which occurs as a result of single-strand cleaving mechanism. As the name implies, it functions optimally at acid pH because it is commonly found in low pH environment of lysosomes.
Exodeoxyribonuclease I is an enzyme that catalyses the following chemical reaction:

The PDE2 enzyme is one of 21 different phosphodiesterases (PDE) found in mammals. These different PDEs can be subdivided to 11 families. The different PDEs of the same family are functionally related despite the fact that their amino acid sequences show considerable divergence. The PDEs have different substrate specificities. Some are cAMP selective hydrolases, others are cGMP selective hydrolases and the rest can hydrolyse both cAMP and cGMP.
Flap endonucleases are a class of nucleolytic enzymes that act as both 5'-3' exonucleases and structure-specific endonucleases on specialised DNA structures that occur during the biological processes of DNA replication, DNA repair, and DNA recombination. Flap endonucleases have been identified in eukaryotes, prokaryotes, archaea, and some viruses. Organisms can have more than one FEN homologue; this redundancy may give an indication of the importance of these enzymes. In prokaryotes, the FEN enzyme is found as an N-terminal domain of DNA polymerase I, but some prokaryotes appear to encode a second homologue.
Deoxyribonuclease IV (phage-T4-induced) is catalyzes the degradation nucleotides in DsDNA by attacking the 5'-terminal end.

T7 DNA polymerase is an enzyme used during the DNA replication of the T7 bacteriophage. During this process, the DNA polymerase “reads” existing DNA strands and creates two new strands that match the existing ones. The T7 DNA polymerase requires a host factor, E. coli thioredoxin, in order to carry out its function. This helps stabilize the binding of the necessary protein to the primer-template to improve processivity by more than 100-fold, which is a feature unique to this enzyme. It is a member of the Family A DNA polymerases, which include E. coli DNA polymerase I and Taq DNA polymerase.

Venom exonuclease is an enzyme. This enzyme catalyses the following chemical reaction

Ribonuclease T is a ribonuclease enzyme involved in the maturation of transfer RNA and ribosomal RNA in bacteria, as well as in DNA repair pathways. It is a member of the DnaQ family of exonucleases and non-processively acts on the 3' end of single-stranded nucleic acids. RNase T is capable of cleaving both DNA and RNA, with extreme sequence specificity discriminating against cytosine at the 3' end of the substrate.

Phospholipase D3, also known as PLD3, is a protein that in humans is encoded by the PLD3 gene. PLD3 belongs to the phospholipase D superfamily because it contains the two HKD motifs common to members of the phospholipase D family, however, it has no known catalytic function similar to PLD1 or PLD2. PLD3 serves as a ssDNA 5' exonuclease in antigen presenting cells. PLD3 is highly expressed in the brain in both humans and mice, and is mainly localized in the endoplasmic reticulum (ER) and the lysosome.

Mitochondrial genome maintenance exonuclease 1, abbreviated as MGME1, is an enzyme that in humans is encoded by the MGME1 gene. MGME1 is a 344 amino acids long protein belonging to the PD-(D/E)XK family of nucleases. It localizes to mitochondria where it is important for maintenance of the mitochondrial genome. Loss of function mutations in MGME1 lead to defects in mitochondrial DNA, including mitochondrial DNA depletion, duplications, deletions and increased replication intermediates. Also, there is an accumulation of 7S DNA, a short single stranded linear DNA strand. MGME1 deficiency in humans leads to multisystemic mitochondrial disease.













