Related Research Articles

Pseudomonadota is a major phylum of Gram-negative bacteria. Currently, they are considered the predominant phylum within the realm of bacteria. They are naturally found as pathogenic and free-living (non-parasitic) genera. The phylum comprises six classes Acidithiobacillia, Alphaproteobacteria, Betaproteobacteria, Gammaproteobacteria, Hydrogenophilia, and Zetaproteobacteria. The Pseudomonadota are widely diverse, with differences in morphology, metabolic processes, relevance to humans, and ecological influence.
The Aquificota phylum is a diverse collection of bacteria that live in harsh environmental settings. The name Aquificota was given to this phylum based on an early genus identified within this group, Aquifex, which is able to produce water by oxidizing hydrogen. They have been found in springs, pools, and oceans. They are autotrophs, and are the primary carbon fixers in their environments. These bacteria are Gram-negative, non-spore-forming rods. They are true bacteria as opposed to the other inhabitants of extreme environments, the Archaea.
Fibrobacterota is a small bacterial phylum which includes many of the major rumen bacteria, allowing for the degradation of plant-based cellulose in ruminant animals. Members of this phylum were categorized in other phyla. The genus Fibrobacter was removed from the genus Bacteroides in 1988.

Gracilicutes is a clade in bacterial phylogeny.

Terrabacteria is a taxon containing approximately two-thirds of prokaryote species, including those in the gram positive phyla as well as the phyla "Cyanobacteria", Chloroflexota, and Deinococcota.
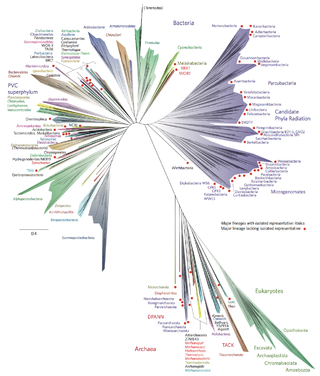
Bacterial phyla constitute the major lineages of the domain Bacteria. While the exact definition of a bacterial phylum is debated, a popular definition is that a bacterial phylum is a monophyletic lineage of bacteria whose 16S rRNA genes share a pairwise sequence identity of ~75% or less with those of the members of other bacterial phyla.

Bacterial taxonomy is subfield of taxonomy devoted to the classification of bacteria specimens into taxonomic ranks.
There are several models of the Branching order of bacterial phyla, one of these was proposed in 1987 paper by Carl Woese.
There are several models of the Branching order of bacterial phyla, the most cited of these was proposed in 1987 paper by Carl Woese. This cladogram was later expanded by Rappé and Giovanoni in 2003 to include newly discovered phyla. Clear names are added in parentheses, see list of bacterial phyla.
There are several models of the Branching order of bacterial phyla, one of these was proposed in 2006 by Ciccarelli et al. for their iTOL project.
There are several models of the Branching order of bacterial phyla, one of these was proposed in 2001 by Gupta based on conserved indels or protein, termed "protein signatures", an alternative approach to molecular phylogeny. Some problematic exceptions and conflicts are present to these conserved indels, however, they are in agreement with several groupings of classes and phyla. One feature of the cladogram obtained with this method is the clustering of cell wall morphology from monoderms to transitional diderms to traditional diderms.
There are several models of the Branching order of bacterial phyla, one of these was proposed in 2002 and 2004 by Thomas Cavalier-Smith. In this frame of work, the branching order of the major lineage of bacteria are determined based on some morphological characters, such as cell wall structure, and not based on the molecular evidence.

'The All-Species Living Tree' Project is a collaboration between various academic groups/institutes, such as ARB, SILVA rRNA database project, and LPSN, with the aim of assembling a database of 16S rRNA sequences of all validly published species of Bacteria and Archaea. At one stage, 23S sequences were also collected, but this has since stopped.

Prokaryotic ubiquitin-like protein (Pup) is a functional analog of ubiquitin found in the prokaryote Mycobacterium tuberculosis. Like ubiquitin, Pup serves to direct proteins to the proteasome for degradation in the Pup-proteasome system (PPS). However, the enzymology of ubiquitylation and pupylation is different, owing to their distinct evolutionary origins. In contrast to the three-step reaction of ubiquitylation, pupylation requires only two steps, and thus only two enzymes are involved in pupylation. The enzymes involved in pupylation are descended from glutamine synthetase.

Hydrobacteria is a taxon containing approximately one-third of prokaryote species, mostly gram-negative bacteria and their relatives. It was found to be the closest relative of an even larger group of Bacteria, Terrabacteria, which are mostly gram-positive bacteria. The name Hydrobacteria refers to the moist environment inferred for the common ancestor of those species. In contrast, species of Terrabacteria possess adaptations for life on land.

Stephen Blair Hedges is Laura H. Carnell Professor of Science and director of the Center for Biodiversity at Temple University where he researches the tree of life and leads conservation efforts in Haiti and elsewhere. He co-founded Haiti National Trust.
Bacterial evolution may refer to the biological evolution of bacteria as studied in:

The evolution of bacteria has progressed over billions of years since the Precambrian time with their first major divergence from the archaeal/eukaryotic lineage roughly 3.2-3.5 billion years ago. This was discovered through gene sequencing of bacterial nucleoids to reconstruct their phylogeny. Furthermore, evidence of permineralized microfossils of early prokaryotes was also discovered in the Australian Apex Chert rocks, dating back roughly 3.5 billion years ago during the time period known as the Precambrian time. This suggests that an organism in of the phylum Thermotogota was the most recent common ancestor of modern bacteria.
The candidate phyla radiation is a large evolutionary radiation of bacterial lineages whose members are mostly uncultivated and only known from metagenomics and single cell sequencing. They have been described as nanobacteria or ultra-small bacteria due to their reduced size (nanometric) compared to other bacteria.
References
- ↑ Battistuzzi, F. U.; Hedges, S. B. (2008). "A Major Clade of Prokaryotes with Ancient Adaptations to Life on Land". Molecular Biology and Evolution. 26 (2): 335–343. doi: 10.1093/molbev/msn247 . PMID 18988685.
- ↑ Battistuzzi, F. U.; Feijao, A.; Hedges, S. B. (2004). "A genomic timescale of prokaryote evolution: insights into the origin of methanogenesis, phototrophy, and the colonization of land". BMC Evolutionary Biology. 4: 44. doi: 10.1186/1471-2148-4-44 . PMC 533871 . PMID 15535883.