Related Research Articles

Pseudomonadota is a major phylum of Gram-negative bacteria. Currently, they are considered the predominant phylum within the realm of bacteria. They are naturally found as pathogenic and free-living (non-parasitic) genera. The phylum comprises six classes Acidithiobacillia, Alphaproteobacteria, Betaproteobacteria, Gammaproteobacteria, Hydrogenophilia, and Zetaproteobacteria. The Pseudomonadota are widely diverse, with differences in morphology, metabolic processes, relevance to humans, and ecological influence.
Division is a taxonomic rank in biological classification that is used differently in zoology and in botany.

In biological taxonomy, a domain, also dominion, superkingdom, realm, or empire, is the highest taxonomic rank of all organisms taken together. It was introduced in the three-domain system of taxonomy devised by Carl Woese, Otto Kandler and Mark Wheelis in 1990.

Bacillota is a phylum of bacteria, most of which have gram-positive cell wall structure. The renaming of phyla such as Firmicutes in 2021 remains controversial among microbiologists, many of whom continue to use the earlier names of long standing in the literature.
The Aquificota phylum is a diverse collection of bacteria that live in harsh environmental settings. The name Aquificota was given to this phylum based on an early genus identified within this group, Aquifex, which is able to produce water by oxidizing hydrogen. They have been found in springs, pools, and oceans. They are autotrophs, and are the primary carbon fixers in their environments. These bacteria are Gram-negative, non-spore-forming rods. They are true bacteria as opposed to the other inhabitants of extreme environments, the Archaea.
The Thermotogota are a phylum of the domain Bacteria. The phylum contains a single class, Thermotogae. The phylum Thermotogota is composed of Gram-negative staining, anaerobic, and mostly thermophilic and hyperthermophilic bacteria.

Gracilicutes is a clade in bacterial phylogeny.
The Chloroflexota are a phylum of bacteria containing isolates with a diversity of phenotypes, including members that are aerobic thermophiles, which use oxygen and grow well in high temperatures; anoxygenic phototrophs, which use light for photosynthesis ; and anaerobic halorespirers, which uses halogenated organics as electron acceptors.
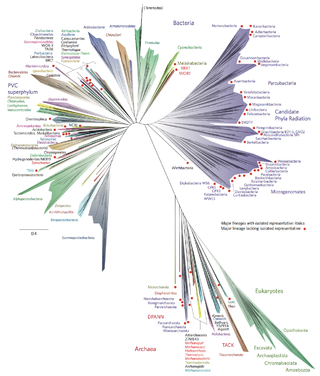
Bacterial phyla constitute the major lineages of the domain Bacteria. While the exact definition of a bacterial phylum is debated, a popular definition is that a bacterial phylum is a monophyletic lineage of bacteria whose 16S rRNA genes share a pairwise sequence identity of ~75% or less with those of the members of other bacterial phyla.

Bacterial taxonomy is subfield of taxonomy devoted to the classification of bacteria specimens into taxonomic ranks.
There are several models of the Branching order of bacterial phyla, one of these was proposed in 1987 paper by Carl Woese.
There are several models of the Branching order of bacterial phyla, the most cited of these was proposed in 1987 paper by Carl Woese. This cladogram was later expanded by Rappé and Giovanoni in 2003 to include newly discovered phyla. Clear names are added in parentheses, see list of bacterial phyla.
There are several models of the Branching order of bacterial phyla, one of these was proposed in 2004 by Battistuzzi and Hedges, note the coinage of the taxa Terrabacteria and Hydrobacteria.
There are several models of the Branching order of bacterial phyla, one of these was proposed in 2001 by Gupta based on conserved indels or protein, termed "protein signatures", an alternative approach to molecular phylogeny. Some problematic exceptions and conflicts are present to these conserved indels, however, they are in agreement with several groupings of classes and phyla. One feature of the cladogram obtained with this method is the clustering of cell wall morphology from monoderms to transitional diderms to traditional diderms.
There are several models of the Branching order of bacterial phyla, one of these was proposed in 2002 and 2004 by Thomas Cavalier-Smith. In this frame of work, the branching order of the major lineage of bacteria are determined based on some morphological characters, such as cell wall structure, and not based on the molecular evidence.

'The All-Species Living Tree' Project is a collaboration between various academic groups/institutes, such as ARB, SILVA rRNA database project, and LPSN, with the aim of assembling a database of 16S rRNA sequences of all validly published species of Bacteria and Archaea. At one stage, 23S sequences were also collected, but this has since stopped.
Bacterial evolution may refer to the biological evolution of bacteria as studied in:
There are several models of the branching order of bacterial phyla, one of these is the Genome Taxonomy Database (GTDB).
Branching order of bacterial phyla may refer to these models:
References
- ↑ Ciccarelli, F. D.; Doerks, T; Von Mering, C; Creevey, CJ; Snel, B; Bork, P (2006). "Toward Automatic Reconstruction of a Highly Resolved Tree of Life". Science. 311 (5765): 1283–1287. Bibcode:2006Sci...311.1283C. CiteSeerX 10.1.1.381.9514 . doi:10.1126/science.1123061. PMID 16513982.