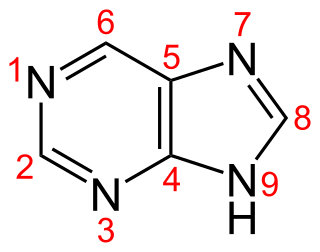
Purine is a heterocyclic aromatic organic compound that consists of two rings fused together. It is water-soluble. Purine also gives its name to the wider class of molecules, purines, which include substituted purines and their tautomers. They are the most widely occurring nitrogen-containing heterocycles in nature.
A salvage pathway is a pathway in which a biological product is produced from intermediates in the degradative pathway of its own or a similar substance. The term often refers to nucleotide salvage in particular, in which nucleotides are synthesized from intermediates in their degradative pathway.
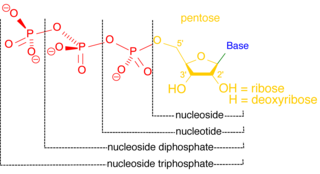
A nucleoside triphosphate is a nucleoside containing a nitrogenous base bound to a 5-carbon sugar, with three phosphate groups bound to the sugar. They are the molecular precursors of both DNA and RNA, which are chains of nucleotides made through the processes of DNA replication and transcription. Nucleoside triphosphates also serve as a source of energy for cellular reactions and are involved in signalling pathways.

Inosinic acid or inosine monophosphate (IMP) is a nucleotide. Widely used as a flavor enhancer, it is typically obtained from chicken byproducts or other meat industry waste. Inosinic acid is important in metabolism. It is the ribonucleotide of hypoxanthine and the first nucleotide formed during the synthesis of purine nucleotides. It can also be formed by the deamination of adenosine monophosphate by AMP deaminase. It can be hydrolysed to inosine.

Nucleic acid metabolism is a collective term that refers to the variety of chemical reactions by which nucleic acids are either synthesized or degraded. Nucleic acids are polymers made up of a variety of monomers called nucleotides. Nucleotide synthesis is an anabolic mechanism generally involving the chemical reaction of phosphate, pentose sugar, and a nitrogenous base. Degradation of nucleic acids is a catabolic reaction and the resulting parts of the nucleotides or nucleobases can be salvaged to recreate new nucleotides. Both synthesis and degradation reactions require multiple enzymes to facilitate the event. Defects or deficiencies in these enzymes can lead to a variety of diseases.
Purine metabolism refers to the metabolic pathways to synthesize and break down purines that are present in many organisms.

Xanthosine monophosphate (xanthylate) is an intermediate in purine metabolism. It is a ribonucleoside monophosphate. It is formed from IMP via the action of IMP dehydrogenase, and it forms GMP via the action of GMP synthase. Also, XMP can be released from XTP by enzyme deoxyribonucleoside triphosphate pyrophosphohydrolase containing (d)XTPase activity.

Guanosine monophosphate synthetase, also known as GMPS is an enzyme that converts xanthosine monophosphate to guanosine monophosphate.

Xanthosine 5'-triphosphate (XTP) is a nucleotide that is not produced by - and has no known function in - living cells. Uses of XTP are, in general, limited to experimental procedures on enzymes that bind other nucleotides. Deamination of purine bases can result in accumulation of such nucleotides as ITP, dITP, XTP, and dXTP.
In enzymology, an ATP diphosphatase (EC 3.6.1.8) is an enzyme that catalyzes the chemical reaction
In enzymology, a dCTP diphosphatase (EC 3.6.1.12) is an enzyme that catalyzes the chemical reaction

In Enzymology, a dUTP diphosphatase (EC 3.6.1.23) is an enzyme that catalyzes the chemical reaction
In enzymology, a guanosine-5'-triphosphate,3'-diphosphate diphosphatase (EC 3.6.1.40) is an enzyme that catalyzes the chemical reaction

In enzymology, a nucleoside-diphosphatase (EC 3.6.1.6) is an enzyme that catalyzes the chemical reaction
In enzymology, a nucleoside-triphosphatase(NTPase) (EC 3.6.1.15) is an enzyme that catalyzes the chemical reaction
In enzymology, a nucleotide diphosphatase (EC 3.6.1.9) is an enzyme that catalyzes the chemical reaction
In enzymology, a phosphoribosyl-ATP diphosphatase (EC 3.6.1.31) is an enzyme that catalyzes the chemical reaction

Inosine triphosphate pyrophosphatase is an enzyme that in humans is encoded by the ITPA gene, by the rdgB gene in bacteria E.coli and the HAM1 gene in yeast S. cerevisiae; the protein is also encoded by some RNA viruses of the Potyviridae family. Two transcript variants encoding two different isoforms have been found for this gene. Also, at least two other transcript variants have been identified which are probably regulatory rather than protein-coding.
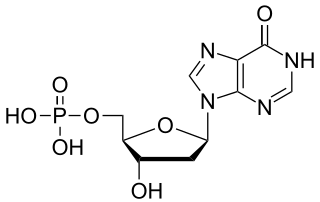
Deoxyinosine monophosphate (dIMP) is a nucleoside monophosphate and a derivative of inosinic acid. It can be formed by the deamination of the purine base in deoxyadenosine monophosphate (dAMP). The enzyme deoxyribonucleoside triphosphate pyrophosphohydrolase, encoded by YJR069C in S. cerevisiae and containing (d)ITPase and (d)XTPase activities, hydrolyses dITP, resulting in the release of pyrophosphate and dIMP.

Inosine triphosphate (ITP) is an intermediate in the purine metabolism pathway, seen in the synthesis of ATP and GTP. It comprises an inosine nucleotide containing three phosphate groups esterified to the sugar moiety.













