Tryptophan synthase or tryptophan synthetase is an enzyme that catalyzes the final two steps in the biosynthesis of tryptophan. It is commonly found in Eubacteria, Archaebacteria, Protista, Fungi, and Plantae. However, it is absent from Animalia. It is typically found as an α2β2 tetramer. The α subunits catalyze the reversible formation of indole and glyceraldehyde-3-phosphate (G3P) from indole-3-glycerol phosphate (IGP). The β subunits catalyze the irreversible condensation of indole and serine to form tryptophan in a pyridoxal phosphate (PLP) dependent reaction. Each α active site is connected to a β active site by a 25 Ångstrom long hydrophobic channel contained within the enzyme. This facilitates the diffusion of indole formed at α active sites directly to β active sites in a process known as substrate channeling. The active sites of tryptophan synthase are allosterically coupled.

A catalytic triad is a set of three coordinated amino acids that can be found in the active site of some enzymes. Catalytic triads are most commonly found in hydrolase and transferase enzymes. An acid-base-nucleophile triad is a common motif for generating a nucleophilic residue for covalent catalysis. The residues form a charge-relay network to polarise and activate the nucleophile, which attacks the substrate, forming a covalent intermediate which is then hydrolysed to release the product and regenerate free enzyme. The nucleophile is most commonly a serine or cysteine amino acid, but occasionally threonine or even selenocysteine. The 3D structure of the enzyme brings together the triad residues in a precise orientation, even though they may be far apart in the sequence.

Indole alkaloids are a class of alkaloids containing a structural moiety of indole; many indole alkaloids also include isoprene groups and are thus called terpene indole or secologanin tryptamine alkaloids. Containing more than 4100 known different compounds, it is one of the largest classes of alkaloids. Many of them possess significant physiological activity and some of them are used in medicine. The amino acid tryptophan is the biochemical precursor of indole alkaloids.

α-Glucosidase (EC 3.2.1.20, is a glucosidase located in the brush border of the small intestine that acts upon α bonds:
Ajmaline is an alkaloid that is classified as a 1-A antiarrhythmic agent. It is often used to induce arrhythmic contraction in patients suspected of having Brugada syndrome. Individuals suffering from Brugada syndrome will be more susceptible to the arrhythmogenic effects of the drug, and this can be observed on an electrocardiogram as an ST elevation.
In enzymology, a vellosimine dehydrogenase (EC 1.1.1.273) is an enzyme that catalyzes the chemical reaction
Strictosidine synthase (EC 4.3.3.2) is an enzyme in alkaloid biosynthesis that catalyses the condensation of tryptamine with secologanin to form strictosidine in a formal Pictet–Spengler reaction:

The enzyme acetylxylan esterase catalyzes the deacetylation of xylans and xylo-oligosaccharides.
The enzyme α-amino-acid esterase (EC 3.1.1.43) catalyzes the reaction
The enzyme arylesterase (EC 3.1.1.2) catalyzes the reaction
The enzyme bis(2-ethylhexyl)phthalate esterase (EC 3.1.1.60) catalyzes the reaction
The enzyme carboxylesterase (or carboxylic-ester hydrolase, EC 3.1.1.1; systematic name carboxylic-ester hydrolase) catalyzes reactions of the following form:

The enzyme cutinase is a member of the hydrolase family. It catalyzes the following reaction:
The enzyme sterol esterase (EC 3.1.1.13) catalyzes the reaction
The enzyme tannase (EC 3.1.1.20) catalyzes the following reaction:
Asymmetric ester hydrolysis with pig liver esterase is the enantioselective conversion of an ester to a carboxylic acid through the action of the enzyme pig liver esterase. Asymmetric ester hydrolysis involves the selective reaction of one of a pair of either enantiotopic or enantiomorphic ester groups.
The enzyme acetylajmaline esterase (EC 3.1.1.80, AAE, 2β(R)-17-O-acetylajmalan:acetylesterase, acetylajmalan esterase; systematic name 17-O-acetylajmaline O-acetylhydrolase) catalyses the following reactions:
3α(S)-strictosidine β-glucosidase (EC 3.2.1.105) is an enzyme with systematic name strictosidine β-D-glucohydrolase. It catalyses the following chemical reaction:
(S)-hydroxynitrile lyase (EC 4.1.2.47, (S)-cyanohydrin producing hydroxynitrile lyase, (S)-oxynitrilase, (S)-HbHNL, (S)-MeHNL, hydroxynitrile lyase, oxynitrilase, HbHNL, MeHNL, (S)-selective hydroxynitrile lyase, (S)-cyanohydrin carbonyl-lyase (cyanide forming), hydroxynitrilase) is an enzyme with systematic name (S)-cyanohydrin lyase (cyanide forming). This enzyme catalyses the interconversion between cyanohydrins and the carbonyl compounds derived from the cyanohydrin with free cyanide, as in the following two chemical reactions:
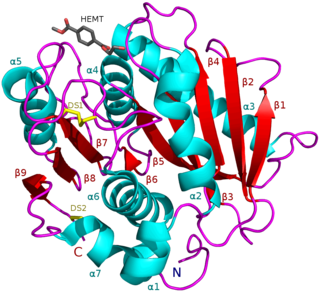
PETases are an esterase class of enzymes that catalyze the breakdown (via hydrolysis) of polyethylene terephthalate (PET) plastic to monomeric mono-2-hydroxyethyl terephthalate (MHET). The idealized chemical reaction is:















