Related Research Articles
A DNA virus is a virus that has DNA as its genetic material and replicates using a DNA-dependent DNA polymerase. The nucleic acid is usually double-stranded DNA (dsDNA) but may also be single-stranded DNA (ssDNA). DNA viruses belong to either Group I or Group II of the Baltimore classification system for viruses. Single-stranded DNA is usually expanded to double-stranded in infected cells. Although Group VII viruses such as hepatitis B contain a DNA genome, they are not considered DNA viruses according to the Baltimore classification, but rather reverse transcribing viruses because they replicate through an RNA intermediate. Notable diseases like smallpox, herpes, and the chickenpox are caused by such DNA viruses.

Mimivirus is a genus of viruses, in the family Mimiviridae. Amoeba serve as their natural hosts. This genus contains a single identified species named Acanthamoeba polyphaga mimivirus (APMV), which serves as its type species. It also refers to a group of phylogenetically related large viruses.
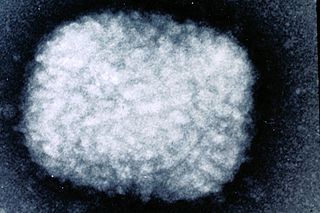
Poxviridae is a family of viruses. Humans, vertebrates, and arthropods serve as natural hosts. There are currently 69 species in this family, divided among 28 genera, which are divided into two subfamilies. Diseases associated with this family include smallpox.
Phycodnaviridae is a family of large (100–560 kb) double-stranded DNA viruses that infect marine or freshwater eukaryotic algae. Viruses within this family have a similar morphology, with an icosahedral capsid. As of 2014, there were 33 species in this family, divided among 6 genera. This family belongs to a super-group of large viruses known as nucleocytoplasmic large DNA viruses. Evidence was published in 2014 suggesting that specific strains of Phycodnaviridae might infect humans rather than just algal species, as was previously believed. Most genera under this family enter the host cell by cell receptor endocytosis and replicate in the nucleus. Phycodnaviridae play important ecological roles by regulating the growth and productivity of their algal hosts. Algal species such Heterosigma akashiwo and the genus Chrysochromulina can form dense blooms which can be damaging to fisheries, resulting in losses in the aquaculture industry. Heterosigma akashiwo virus (HaV) has been suggested for use as a microbial agent to prevent the recurrence of toxic red tides produced by this algal species. Phycodnaviridae cause death and lysis of freshwater and marine algal species, liberating organic carbon, nitrogen and phosphorus into the water, providing nutrients for the microbial loop.

Iridoviridae is a family of viruses with double-stranded DNA genomes. Amphibians, fish, and invertebrates such as arthropods serve as natural hosts. There are currently 12 species in this family, divided among two subfamilies and five genera.
Ascoviridae is a family of double strand DNA viruses that infect primarily invertebrates, mainly noctuids and spodoptera species; it contains two genera, Ascovirus, which contains three species, and Toursvirus with a single species Diadromus pulchellus toursvirus. The type species of Ascovirus is Spodoptera frugiperda ascovirus 1a, which infects the army worm.

Mimiviridae is a family of viruses. Amoeba and other protists serve as natural hosts. The family is divided in up to 4 subfamilies. Viruses in this family belong to the nucleocytoplasmic large DNA virus clade (NCLDV), also referred to as giant viruses.
Mamavirus is a large and complex virus in the Group I family mimiviridae. The virus is exceptionally large, and larger than many bacteria. Mamavirus and other mimiviridae belong to nucleocytoplasmic large DNA virus (NCLDVs) family. Mamavirus can be compared to the similar complex virus mimivirus; mamavirus was so named because it is similar to but larger than mimivirus.
Marseillevirus is a genus of viruses, in the family Marseilleviridae. There are currently only two species in this genus, including the type species Marseillevirus marseillevirus. It is the prototype of a family of nucleocytoplasmic large DNA viruses (NCLDV) of eukaryotes. It was isolated from amoeba.
Cafeteria roenbergensis virus (CroV) is a giant virus that infects the marine bicosoecid flagellate Cafeteria roenbergensis, a member of the microzooplankton community.
Organic Lake is a lake in the Vestfold Hills in eastern Antarctica. It was formed 6,000 years ago when sea levels were higher; it is isolated, rather shallow (7.5m), meromictic, a few hundred meters in diameter and has extremely salty water. It has the highest recorded concentration of dimethyl sulfide in any natural body of water.
Lymphocystivirus is a genus of viruses, in the family Iridoviridae. Fish serve as natural hosts. There is currently only one species in this genus: the type species Lymphocystis disease virus 1. Diseases associated with this genus include: tumor-like growths on the skin.

A giant virus, also known as a girus, is a very large virus, some of which are larger than typical bacteria. They are giant nucleocytoplasmic large DNA viruses (NCLDVs) that have extremely large genomes compared to other viruses and contain many unique genes not found in other life forms.
Marseilleviridae is a family of viruses first named in 2012. The genomes of these viruses are double-stranded DNA. Amoeba are often hosts, but there is evidence that they are found in humans as well. As of 2016, the International Committee on Taxonomy of Viruses recognize four species in this family, divided among 2 genera. It is a member of the nucleocytoplasmic large DNA viruses clade.

Megavirus is a viral genus containing a single identified species named Megavirus chilensis, phylogenetically related to Acanthamoeba polyphaga Mimivirus (APMV). In colloquial speech, Megavirus chilensis is more commonly referred to as just “Megavirus”. Until the discovery of pandoraviruses in 2013, it had the largest capsid diameter of all known viruses, as well as the largest and most complex genome among all known viruses.
Organic Lake virophage (OLV) is a double-stranded DNA virophage. It was detected metagenomically in samples from Organic Lake, Antarctica.
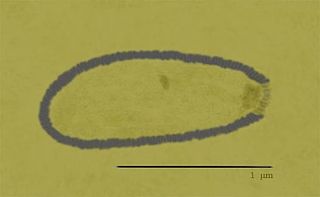
Pithovirus, first described in a 2014 paper, is a genus of giant virus known from one species, Pithovirus sibericum, which infects amoebas. It is a double-stranded DNA virus, and is a member of the nucleocytoplasmic large DNA viruses clade. The 2014 discovery was made when a viable specimen was found in a 30,000-year-old ice core harvested from permafrost in Siberia, Russia.
Chlorovirus, also known as Chlorella virus, is a genus of giant double-stranded DNA viruses, in the family Phycodnaviridae. This genus is found globally in freshwater environments where freshwater microscopic algae serve as natural hosts. There are currently 19 species in this genus including the type species Paramecium bursaria Chlorella virus 1.

Mimivirus-dependent virus Zamilon, or Zamilon, is a virophage, a group of small DNA viruses that infect protists and require a helper virus to replicate; they are a type of satellite virus. Discovered in 2013 in Tunisia, infecting Acanthamoeba polyphaga amoebae, Zamilon most closely resembles Sputnik, the first virophage to be discovered. The name is Arabic for "the neighbour". Its spherical particle is 50–60 nm in diameter, and contains a circular double-stranded DNA genome of around 17 kb, which is predicted to encode 20 polypeptides. A related strain, Zamilon 2, has been identified in North America.

Chrysochromulina ericina virus 01B, or simply Chrysochromulina ericina virus (CeV) is a giant virus in the family Mimiviridae infecting Haptolina ericina, a marine microalgae member of the Haptophyta. CeV is a dsDNA virus.
References
- ↑ Colson P, de Lamballerie X, Fournous G, Raoult D (2012). "Reclassification of giant viruses composing a fourth domain of life in the new order Megavirales". Intervirology. 55 (5): 321–332. doi:10.1159/000336562. PMID 22508375.
- ↑ Colson P, De Lamballerie X, Yutin N, Asgari S, Bigot Y, Bideshi DK, Cheng XW, Federici BA, Van Etten JL, Koonin EV, La Scola B, Raoult D (2013). ""Megavirales", a proposed new order for eukaryotic nucleocytoplasmic large DNA viruses". Arch Virol. 158 (12): 2517–21. doi:10.1007/s00705-013-1768-6. PMC 4066373 . PMID 23812617.
- ↑ Iyer, L. M.; Aravind, L.; Koonin, E. V. (December 2001). "Common Origin of Four Diverse Families of Large Eukaryotic DNA Viruses". Journal of Virology. 75 (23): 11720–34. doi:10.1128/JVI.75.23.11720-11734.2001. PMC 114758 . PMID 11689653.
- ↑ "Ascoviridae—Ascoviridae—dsDNA Viruses—International Committee on Taxonomy of Viruses (ICTV)". International Committee on Taxonomy of Viruses (ICTV). Retrieved 2017-12-07.
- ↑ Asgari, Sassan; Bideshi, Dennis K; Bigot, Yves; Federici, Brian A; Cheng, Xiao-Wen (2017). "ICTV Virus Taxonomy Profile: Ascoviridae". The Journal of General Virology. 98 (1): 4–5. doi:10.1099/jgv.0.000677. ISSN 0022-1317. PMC 5370392 . PMID 28218573.
- ↑ "African swine fever (ASF) | animal disease". Encyclopedia Britannica. Retrieved 2017-12-07.
- ↑ "Iridoviridae—Iridoviridae—dsDNA Viruses—International Committee on Taxonomy of Viruses (ICTV)". International Committee on Taxonomy of Viruses (ICTV). Retrieved 2017-12-07.
- ↑ Boyer, Mickaël; Yutin, Natalya; Pagnier, Isabelle; Barrassi, Lina; Fournous, Ghislain; Espinosa, Leon; Robert, Catherine; Azza, Saïd; Sun, Siyang (2009-12-22). "Giant Marseillevirus highlights the role of amoebae as a melting pot in emergence of chimeric microorganisms". Proceedings of the National Academy of Sciences of the United States of America. 106 (51): 21848–21853. Bibcode:2009PNAS..10621848B. doi:10.1073/pnas.0911354106. ISSN 0027-8424. PMC 2799887 . PMID 20007369.
- ↑ Aherfi, Sarah (2014-10-01). "The expanding family Marseilleviridae". Virology. 466–467: 27–37. doi:10.1016/j.virol.2014.07.014. ISSN 0042-6822. PMID 25104553.
- ↑ Arslan, Defne; Legendre, Matthieu; Seltzer, Virginie; Abergel, Chantal; Claverie, Jean-Michel (2011-10-18). "Distant Mimivirus relative with a larger genome highlights the fundamental features of Megaviridae". Proceedings of the National Academy of Sciences. 108 (42): 17486–17491. Bibcode:2011PNAS..10817486A. doi:10.1073/pnas.1110889108. ISSN 0027-8424. PMC 3198346 . PMID 21987820.
- ↑ Schulz, Frederik; Yutin, Natalya; Ivanova, Natalia N.; Ortega, Davi R.; Lee, Tae Kwon; Vierheilig, Julia; Daims, Holger; Horn, Matthias; Wagner, Michael (2017-04-07). "Giant viruses with an expanded complement of translation system components" (PDF). Science. 356 (6333): 82–85. Bibcode:2017Sci...356...82S. doi:10.1126/science.aal4657. ISSN 0036-8075. PMID 28386012., UCPMS ID: 1889607, PDF
- ↑ Eugene V Koonin, Mart Krupovic, Natalya Yutin: Evolution of double-stranded DNA viruses of eukaryotes: From bacteriophages to transposons to giant viruses, in: ResearchGate Literature Review February 2015, doi: 10.1111/nyas.12728, Figure 3
- ↑ Natalya Yutin et al.: Mimiviridae: clusters of orthologous genes, reconstruction of gene repertoire evolution and proposed expansion of the giant virus family, in: Virol J. 2013; 10: 106, doi: 10.1186/1743-422X-10-106
- ↑ Blog of Carolina Reyes, Kenneth Stedman: Are Phaeocystis globosa viruses (OLPG) and Organic Lake phycodnavirus a part of the Phycodnaviridae or Mimiviridae?, on ResearchGate, Jan. 8, 2016
- ↑ Fumito Maruyama and Shoko Ueki: Evolution and Phylogeny of Large DNA Viruses, Mimiviridae and Phycodnaviridae Including Newly Characterized Heterosigma akashiwo Virus, in: Front. Microbiol., 30 November 2016, doi: 10.3389/fmicb.2016.01942
- ↑ Weijia Zhang et al.: Four novel algal virus genomes discovered from Yellowstone Lake metagenomes, in: Scientific Reports 5, Article number: 15131 (2015), especially Figure 6, doi: 10.1038/srep15131
- ↑ Yong, Ed (2013). "Giant viruses open Pandora's box". Nature. doi:10.1038/nature.2013.13410.
- ↑ Aherfi, Sarah; Colson, Philippe; La Scola, Bernard; Raoult, Didier (2016-03-22). "Giant Viruses of Amoebas: An Update". Frontiers in Microbiology. 7: 349. doi:10.3389/fmicb.2016.00349. ISSN 1664-302X. PMC 4801854 . PMID 27047465.
- ↑ "Biggest Virus Yet Found, May Be Fourth Domain of Life?". 2013-07-19. Retrieved 2017-12-07.
- ↑ Wilson, W. H.; Van Etten, J. L.; Allen, M. J. (2009). The Phycodnaviridae: The Story of How Tiny Giants Rule the World. Current Topics in Microbiology and Immunology. 328. pp. 1–42. doi:10.1007/978-3-540-68618-7_1. ISBN 978-3-540-68617-0. ISSN 0070-217X. PMC 2908299 . PMID 19216434.
- ↑ Ornes, Stephen (2017-07-31). "Return of the giant zombie virus". Science News for Students. Retrieved 2017-12-07.
- ↑ "Pithovirus". viralzone.expasy.org. Retrieved 2017-12-07.
- ↑ Moss, Bernard (2013). "Poxvirus DNA Replication". Cold Spring Harbor Perspectives in Biology. 5 (9): a010199. doi:10.1101/cshperspect.a010199. ISSN 1943-0264. PMC 3753712 . PMID 23838441.
- ↑ Subramaniam, K (14 January 2020). "A New Family of DNA Viruses Causing Disease in Crustaceans from Diverse Aquatic Biomes". mBio. 11 (1). doi:10.1128/mBio.02938-19. PMC 6960288 . PMID 31937645.
- ↑ Bäckström D, Yutin N, Jørgensen SL, Dharamshi J, Homa F, Zaremba-Niedwiedzka K, Spang A, Wolf YI, Koonin EV, Ettema TJ (2019). "Virus genomes from deep sea sediments expand the ocean megavirome and support independent origins of viral gigantism". mBio. 10 (2): e02497-18. doi:10.1128/mBio.02497-18. PMC 6401483 . PMID 30837339. PDF
- ↑ Guglielmini, Julien; Woo, Anthony C.; Krupovic, Mart; Forterre, Patrick; Gaia, Morgan (2019-09-10). "Diversification of giant and large eukaryotic dsDNA viruses predated the origin of modern eukaryotes". Proceedings of the National Academy of Sciences. 116 (39): 19585–19592. doi:10.1073/pnas.1912006116. ISSN 0027-8424. PMC 6765235 . PMID 31506349.
