
Adenylate cyclase is an enzyme with systematic name ATP diphosphate-lyase . It catalyzes the following reaction:

Nucleotides are organic molecules composed of a nitrogenous base, a pentose sugar and a phosphate. They serve as monomeric units of the nucleic acid polymers – deoxyribonucleic acid (DNA) and ribonucleic acid (RNA), both of which are essential biomolecules within all life-forms on Earth. Nucleotides are obtained in the diet and are also synthesized from common nutrients by the liver.
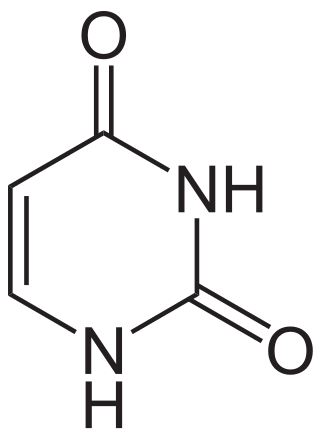
Uracil is one of the four nucleobases in the nucleic acid RNA. The others are adenine (A), cytosine (C), and guanine (G). In RNA, uracil binds to adenine via two hydrogen bonds. In DNA, the uracil nucleobase is replaced by thymine (T). Uracil is a demethylated form of thymine.
Site-directed mutagenesis is a molecular biology method that is used to make specific and intentional mutating changes to the DNA sequence of a gene and any gene products. Also called site-specific mutagenesis or oligonucleotide-directed mutagenesis, it is used for investigating the structure and biological activity of DNA, RNA, and protein molecules, and for protein engineering.

Ribonucleotide reductase (RNR), also known as ribonucleoside diphosphate reductase (rNDP), is an enzyme that catalyzes the formation of deoxyribonucleotides from ribonucleotides. It catalyzes this formation by removing the 2'-hydroxyl group of the ribose ring of nucleoside diphosphates. This reduction produces deoxyribonucleotides. Deoxyribonucleotides in turn are used in the synthesis of DNA. The reaction catalyzed by RNR is strictly conserved in all living organisms. Furthermore, RNR plays a critical role in regulating the total rate of DNA synthesis so that DNA to cell mass is maintained at a constant ratio during cell division and DNA repair. A somewhat unusual feature of the RNR enzyme is that it catalyzes a reaction that proceeds via a free radical mechanism of action. The substrates for RNR are ADP, GDP, CDP and UDP. dTDP is synthesized by another enzyme from dTMP.

Nucleic acid metabolism is a collective term that refers to the variety of chemical reactions by which nucleic acids are either synthesized or degraded. Nucleic acids are polymers made up of a variety of monomers called nucleotides. Nucleotide synthesis is an anabolic mechanism generally involving the chemical reaction of phosphate, pentose sugar, and a nitrogenous base. Degradation of nucleic acids is a catabolic reaction and the resulting parts of the nucleotides or nucleobases can be salvaged to recreate new nucleotides. Both synthesis and degradation reactions require multiple enzymes to facilitate the event. Defects or deficiencies in these enzymes can lead to a variety of diseases.
Pyrophosphatases, also known as diphosphatases, are acid anhydride hydrolases that act upon diphosphate bonds.

Deoxyuridine monophosphate (dUMP), also known as deoxyuridylic acid or deoxyuridylate in its conjugate acid and conjugate base forms, respectively, is a deoxynucleotide.
dnaS or dut is a gene involved in DNA replication in Escherichia coli. It encodes dUTP nucleotidohydrolase, an enzyme responsible for catalyzing the conversion of dUTP to dUMP, thereby ensuring that the organism's DNA contains the nucleobase thymine instead of uracil.

In enzymology, a phenylalanine—tRNA ligase is an enzyme that catalyzes the chemical reaction
In enzymology, a phosphopantothenate—cysteine ligase also known as phosphopantothenoylcysteine synthetase (PPCS) is an enzyme that catalyzes the chemical reaction which constitutes the second of five steps involved in the conversion of pantothenate to Coenzyme A. The reaction is:
In enzymology, a dCTP diphosphatase (EC 3.6.1.12) is an enzyme that catalyzes the chemical reaction

In enzymology, a nucleoside-diphosphatase (EC 3.6.1.6) is an enzyme that catalyzes the chemical reaction
In enzymology, a nucleoside-triphosphatase(NTPase) (EC 3.6.1.15) is an enzyme that catalyzes the chemical reaction
Thiamine-triphosphatase is an enzyme involved in thiamine metabolism. It catalyzes the chemical reaction
In enzymology, a thymidine-triphosphatase (EC 3.6.1.39) is an enzyme that catalyzes the chemical reaction
DUTP pyrophosphatase, also known as DUT, is an enzyme which in humans is encoded by the DUT gene on chromosome 15.

Inosine triphosphate pyrophosphatase is an enzyme that in humans is encoded by the ITPA gene, by the rdgB gene in bacteria E.coli and the HAM1 gene in yeast S. cerevisiae; the protein is also encoded by some RNA viruses of the Potyviridae family. Two transcript variants encoding two different isoforms have been found for this gene. Also, at least two other transcript variants have been identified which are probably regulatory rather than protein-coding.

In molecular biology, the ars operon is an operon found in several bacterial taxon. It is required for the detoxification of arsenate, arsenite, and antimonite. This system transports arsenite and antimonite out of the cell. The pump is composed of two polypeptides, the products of the arsA and arsB genes. This two-subunit enzyme produces resistance to arsenite and antimonite. Arsenate, however, must first be reduced to arsenite before it is extruded. A third gene, arsC, expands the substrate specificity to allow for arsenate pumping and resistance. ArsC is an approximately 150-residue arsenate reductase that uses reduced glutathione (GSH) to convert arsenate to arsenite with a redox active cysteine residue in the active site. ArsC forms an active quaternary complex with GSH, arsenate, and glutaredoxin 1 (Grx1). The three ligands must be present simultaneously for reduction to occur.
In molecular biology, an arginine finger is an amino acid residue of some enzymes. Arginine fingers are often found in the protein superfamily of AAA+ ATPases, GTPases, and dUTPases, where they assist in the catalysis of the gamma phosphate or gamma and beta phosphates from ATP or GTP, which creates a release of energy which can be used to perform cellular work. They are also found in GTPase-activating proteins (GAP). Thus, they are essential for many forms of life, and are highly conserved. Arginine fingers function through non-covalent interactions. They may also assist in dimerization, and while they are found in a wide variety of enzymes, they are not ubiquitous.













