| |
| Names | |
|---|---|
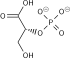
| Preferred IUPAC name (2R)-2-Hydroxy-3-(phosphonooxy)propanoic acid | |
| Identifiers | |
3D model (JSmol) | |
| ChEBI | |
| ChEMBL | |
| ChemSpider | |
| DrugBank | |
| KEGG | |
PubChem CID | |
CompTox Dashboard (EPA) | |
| |
| |
| Properties | |
| C3H7O7P | |
| Molar mass | 186.06 g/mol |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa). | |
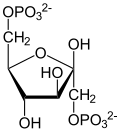
3-Phosphoglyceric acid (3PG, 3-PGA, or PGA) is the conjugate acid of 3-phosphoglycerate or glycerate 3-phosphate (GP or G3P). [1] This glycerate is a biochemically significant metabolic intermediate in both glycolysis and the Calvin-Benson cycle. The anion is often termed as PGA when referring to the Calvin-Benson cycle. In the Calvin-Benson cycle, 3-phosphoglycerate is typically the product of the spontaneous scission of an unstable 6-carbon intermediate formed upon CO2 fixation. Thus, two equivalents of 3-phosphoglycerate are produced for each molecule of CO2 that is fixed. [2] [3] [4] In glycolysis, 3-phosphoglycerate is an intermediate following the dephosphorylation (reduction) of 1,3-bisphosphoglycerate. [4] : 14