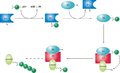
Relationships
A ubiquitin-activating enzyme, or E1, first activates the ubiquitin by covalently attaching the molecule to its active site cysteine residue. The activated ubiquitin is then transferred to an E2 cysteine. Once conjugated to ubiquitin, the E2 molecule binds one of several ubiquitin ligases or E3s via a structurally conserved binding region. The E3 molecule is responsible for binding the target protein substrate and transferring the ubiquitin from the E2 cysteine to a lysine residue on the target protein. [1]
- Schematic diagram of the ubiquitylation system.
A particular cell usually contains only a few types of E1 molecule, a greater diversity of E2s, and a very large variety of E3s. In humans, there are about 30 E2s which can bind with one of the 600+ E3s. [2] The E3 molecules responsible for substrate identification and binding are thus the mechanisms of substrate specificity in proteasomal degradation. Each type of E2 can associate with many E3s. [3]
E2s can also be used to study protein folding mechanisms. Since the ubiquitylation system is shared across all organisms, studies can use modified E2 proteins in order to understand the overall system for how all organisms process proteins. [4] There are also some proteins which can act as both and E2 and an E3 containing domains which cover both E2 and E3 functionality. [5]