Circoviridae is a family of DNA viruses. Birds and mammals serve as natural hosts. There are 101 species in this family, assigned to 2 genera. Diseases associated with this family include: PCV-2: postweaning multisystemic wasting syndrome; CAV: chicken infectious anemia.
Porcine circoviral disease (PCVD), also known as porcine circovirus associated disease (PCVAD), is a disease seen in domestic pigs. This disease causes illness in piglets, with clinical signs including progressive loss of body condition, visibly enlarged lymph nodes, difficulty in breathing, and sometimes diarrhea, pale skin, and jaundice. PCVD is very damaging to the pig-producing industry and has been reported worldwide. PCVD is caused by Porcine circovirus 2 (PCV-2).
Betaarterivirus suid 1, commonly Porcine reproductive and respiratory syndrome virus (PRRSV), is a virus that causes a disease of pigs, called porcine reproductive and respiratory syndrome (PRRS), also known as blue-ear pig disease. This economically important, panzootic disease causes reproductive failure in breeding stock and respiratory tract illness in young pigs.
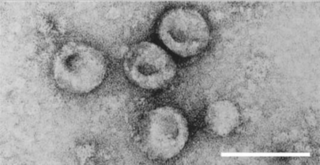
Pestivirus is a genus of viruses, in the family Flaviviridae. Viruses in the genus Pestivirus infect mammals, including members of the family Bovidae and the family Suidae. There are 11 species in this genus. Diseases associated with this genus include: hemorrhagic syndromes, abortion, and fatal mucosal disease.
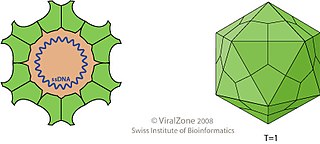
Porcine circovirus (PCV) is a group of four single-stranded DNA viruses that are non-enveloped with an unsegmented circular genome. They are members of the genus Circovirus that can infect pigs. The viral capsid is icosahedral and approximately 17 nm in diameter.
Porcine parvovirus (PPV), a virus in the species Ungulate protoparvovirus 1 of genus Protoparvovirus in the virus family Parvoviridae, causes reproductive failure of swine characterized by embryonic and fetal infection and death, usually in the absence of outward maternal clinical signs. The disease develops mainly when seronegative dams are exposed oronasally to the virus anytime during about the first half of gestation, and conceptuses are subsequently infected transplacentally before they become immunocompetent. There is no definitive evidence that infection of swine other than during gestation is of any clinical or economic significance. The virus is ubiquitous among swine throughout the world and is enzootic in most herds that have been tested. Diagnostic surveys have indicated that PPV is the major infectious cause of embryonic and fetal death. In addition to its direct causal role in reproductive failure, PPV can potentiate the effects of porcine circovirus type II (PCV2) infection in the clinical course of postweaning multisystemic wasting syndrome (PMWS).
Aujeszky's disease, usually called pseudorabies in the United States, is a viral disease in swine that is endemic in most parts of the world. It is caused by Suid herpesvirus 1 (SuHV-1). Aujeszky's disease is considered to be the most economically important viral disease of swine in areas where classical swine fever has been eradicated. Other mammals, such as cattle, sheep, goats, cats, dogs, and raccoons, are also susceptible. The disease is usually fatal in these animal species.

Psittacine beak and feather disease (PBFD) is a viral disease affecting all Old World and New World parrots. The causative virus—beak and feather disease virus (BFDV)—belongs to the taxonomic genus Circovirus, family Circoviridae. It attacks the feather follicles and the beak and claw matrices of the bird, causing progressive feather, claw and beak malformation and necrosis. In later stages of the disease, feather shaft constriction occurs, hampering development until eventually all feather growth stops. It occurs in an acutely fatal form and a chronic form.

Gammaherpesvirinae is a subfamily of viruses in the order Herpesvirales and in the family Herpesviridae. Viruses in Gammaherpesvirinae are distinguished by reproducing at a more variable rate than other subfamilies of Herpesviridae. Mammals serve as natural hosts. There are 43 species in this subfamily, divided among 7 genera with three species unassigned to a genus. Diseases associated with this subfamily include: HHV-4: infectious mononucleosis. HHV-8: Kaposi's sarcoma.

Veterinary virology is the study of viruses in non-human animals. It is an important branch of veterinary medicine.
Gyrovirus is a genus of viruses, in the family Anelloviridae. Until 2011, chicken anemia virus was the only Gyrovirus identified, but since then gyroviruses have also been identified in humans. Diseases associated with this genus include: chicken infectious anemia, which is associated with depletion of cortical thymocytes and erythroblastoid cells.
Alphatorquevirus is a genus of viruses in the family Anelloviridae, in group II in the Baltimore classification. It encompasses numerous species of the virus that was formerly known as TTV, torque teno virus, SENV, SANBAN, and various others. The genus contains 26 species.
Porcine epidemic diarrhea is a condition caused by the porcine epidemic diarrhea virus that leads to severe gastrointestinal disease in pigs.

The bat virome is the group of viruses associated with bats. Bats host a diverse array of viruses, including all seven types described by the Baltimore classification system: (I) double-stranded DNA viruses; (II) single-stranded DNA viruses; (III) double-stranded RNA viruses; (IV) positive-sense single-stranded RNA viruses; (V) negative-sense single-stranded RNA viruses; (VI) positive-sense single-stranded RNA viruses that replicate through a DNA intermediate; and (VII) double-stranded DNA viruses that replicate through a single-stranded RNA intermediate. The greatest share of bat-associated viruses identified as of 2020 are of type IV, in the family Coronaviridae.
Canine circovirus, first isolated in 2012, is a small non-enveloped, icosahedral, single-stranded DNA virus that infects domestic dogs and wild canids exclusively. It is a member of the Circoviridae family and the genus Circovirus. There are currently 11 species of known circoviruses that have been identified to affect a wide variety of birds and mammals. As seen with all extensively studied circoviruses, the diameter ranges between approximately 15 and 25 nanometers. The icosahedral triangulation number is 1, the smallest size a viral capsid can be, in which there are a total of 60 protein subunits that make up the capsid. CaCV is not to be confused with canine coronavirus, another diarrhea-causing agent within the family Coronaviridae, or porcine circoviruses which are a members of the same genus as CaCV but only seen in pigs. CaCV was the first Circovirus to be identified that infects a mammal species other than pigs.
Betaarterivirus suid 2 is a species of enveloped, positive-strand RNA viruses which infect domestic pigs. Members of the species are also known as porcine reproductive and respiratory syndrome virus 2. Member viruses are a type of the porcine reproductive and respiratory syndrome viruses (PRRSV). The two types of PRRSV are distinguished by which genomic cluster they are associated with. Type 1 is associated with a LV cluster. Type 2 is associated with a VR2332 cluster.
Torque teno sus virus, belonging to the family Anelloviridae, is a group of virus strains that are non-enveloped, with a single-stranded circular DNA genome ranging from 2.6 to 2.8 kb in size. These swine infecting anelloviruses are divided into two genera: Iotatorquevirus and Kappatorquevirus. Torque teno sus virus has been found in pigs worldwide. TTSuVs are mainly transmitted by fecal-oral route. The prevalence of these viruses is relatively high. For now, there is not known disease caused exclusively by TTSuV. There is the possibility that TTSuV may worsen the progression of other diseases and therefore increase the economic losses for pig industry.

Redondoviruses are a family of human-associated DNA viruses. Their name derives from the inferred circular structure of the viral genome . Redondoviruses have been identified in DNA sequence based surveys of samples from humans, primarily samples from the oral cavity and upper airway.

Monodnaviria is a realm of viruses that includes all single-stranded DNA viruses that encode an endonuclease of the HUH superfamily that initiates rolling circle replication of the circular viral genome. Viruses descended from such viruses are also included in the realm, including certain linear single-stranded DNA (ssDNA) viruses and circular double-stranded DNA (dsDNA) viruses. These atypical members typically replicate through means other than rolling circle replication.
Xiang-Jin Meng, also known as X.J. Meng, is a Chinese-born American virologist. He is a university distinguished professor at Virginia Tech. He studies emerging, re-emerging and zoonotic viruses of veterinary and human public health significance. He was elected a member of the National Academy of Sciences in 2016, a Fellow of the National Academy of Inventors in 2014, a Fellow of the American Academy of Microbiology in 2012, and a Fellow of the American Association for the Advancement of Science.








