
Computational biology refers to the use of data analysis, mathematical modeling and computational simulations to understand biological systems and relationships. An intersection of computer science, biology, and big data, the field also has foundations in applied mathematics, chemistry, and genetics. It differs from biological computing, a subfield of computer science and engineering which uses bioengineering to build computers.

A generegulatory network (GRN) is a collection of molecular regulators that interact with each other and with other substances in the cell to govern the gene expression levels of mRNA and proteins which, in turn, determine the function of the cell. GRN also play a central role in morphogenesis, the creation of body structures, which in turn is central to evolutionary developmental biology (evo-devo).

In mathematics, computer science and network science, network theory is a part of graph theory. It defines networks as graphs where the vertices or edges possess attributes. Network theory analyses these networks over the symmetric relations or asymmetric relations between their (discrete) components.
In molecular biology, an interactome is the whole set of molecular interactions in a particular cell. The term specifically refers to physical interactions among molecules but can also describe sets of indirect interactions among genes.
Modelling biological systems is a significant task of systems biology and mathematical biology. Computational systems biology aims to develop and use efficient algorithms, data structures, visualization and communication tools with the goal of computer modelling of biological systems. It involves the use of computer simulations of biological systems, including cellular subsystems, to both analyze and visualize the complex connections of these cellular processes.

In graph theory and network analysis, indicators of centrality assign numbers or rankings to nodes within a graph corresponding to their network position. Applications include identifying the most influential person(s) in a social network, key infrastructure nodes in the Internet or urban networks, super-spreaders of disease, and brain networks. Centrality concepts were first developed in social network analysis, and many of the terms used to measure centrality reflect their sociological origin.

Personalized medicine, also referred to as precision medicine, is a medical model that separates people into different groups—with medical decisions, practices, interventions and/or products being tailored to the individual patient based on their predicted response or risk of disease. The terms personalized medicine, precision medicine, stratified medicine and P4 medicine are used interchangeably to describe this concept, though some authors and organizations differentiate between these expressions based on particular nuances. P4 is short for "predictive, preventive, personalized and participatory".

KEGG is a collection of databases dealing with genomes, biological pathways, diseases, drugs, and chemical substances. KEGG is utilized for bioinformatics research and education, including data analysis in genomics, metagenomics, metabolomics and other omics studies, modeling and simulation in systems biology, and translational research in drug development.
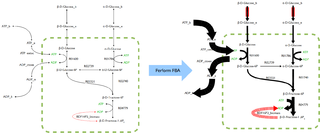
In biochemistry, flux balance analysis (FBA) is a mathematical method for simulating the metabolism of cells or entire unicellular organisms, such as E. coli or yeast, using genome-scale reconstructions of metabolic networks. Genome-scale reconstructions describe all the biochemical reactions in an organism based on its entire genome. These reconstructions model metabolism by focusing on the interactions between metabolites, identifying which metabolites are involved in the various reactions taking place in a cell or organism, and determining the genes that encode the enzymes which catalyze these reactions. In comparison to traditional methods of modeling, FBA is less intensive in terms of the input data required for constructing the model. Simulations performed using FBA are computationally inexpensive and can calculate steady-state metabolic fluxes for large models in a few seconds on modern personal computers. The related method of metabolic pathway analysis seeks to find and list all possible pathways between metabolites.

An enzyme inhibitor is a molecule that binds to an enzyme and blocks its activity. Enzymes are proteins that speed up chemical reactions necessary for life, in which substrate molecules are converted into products. An enzyme facilitates a specific chemical reaction by binding the substrate to its active site, a specialized area on the enzyme that accelerates the most difficult step of the reaction.

Biological network inference is the process of making inferences and predictions about biological networks. By using these networks to analyze patterns in biological systems, such as food-webs, we can visualize the nature and strength of these interactions between species, DNA, proteins, and more.

Modularity is a measure of the structure of networks or graphs which measures the strength of division of a network into modules. Networks with high modularity have dense connections between the nodes within modules but sparse connections between nodes in different modules. Modularity is often used in optimization methods for detecting community structure in networks. Biological networks, including animal brains, exhibit a high degree of modularity. However, modularity maximization is not statistically consistent, and finds communities in its own null model, i.e. fully random graphs, and therefore it cannot be used to find statistically significant community structures in empirical networks. Furthermore, it has been shown that modularity suffers a resolution limit and, therefore, it is unable to detect small communities.
In computational biology, power graph analysis is a method for the analysis and representation of complex networks. Power graph analysis is the computation, analysis and visual representation of a power graph from a graph (networks).

A biological network is a method of representing systems as complex sets of binary interactions or relations between various biological entities. In general, networks or graphs are used to capture relationships between entities or objects. A typical graphing representation consists of a set of nodes connected by edges.

PageRank (PR) is an algorithm used by Google Search to rank web pages in their search engine results. It is named after both the term "web page" and co-founder Larry Page. PageRank is a way of measuring the importance of website pages. According to Google:
PageRank works by counting the number and quality of links to a page to determine a rough estimate of how important the website is. The underlying assumption is that more important websites are likely to receive more links from other websites.
Immunomics is the study of immune system regulation and response to pathogens using genome-wide approaches. With the rise of genomic and proteomic technologies, scientists have been able to visualize biological networks and infer interrelationships between genes and/or proteins; recently, these technologies have been used to help better understand how the immune system functions and how it is regulated. Two thirds of the genome is active in one or more immune cell types and less than 1% of genes are uniquely expressed in a given type of cell. Therefore, it is critical that the expression patterns of these immune cell types be deciphered in the context of a network, and not as an individual, so that their roles be correctly characterized and related to one another. Defects of the immune system such as autoimmune diseases, immunodeficiency, and malignancies can benefit from genomic insights on pathological processes. For example, analyzing the systematic variation of gene expression can relate these patterns with specific diseases and gene networks important for immune functions.

Network controllability concerns the structural controllability of a network. Controllability describes our ability to guide a dynamical system from any initial state to any desired final state in finite time, with a suitable choice of inputs. This definition agrees well with our intuitive notion of control. The controllability of general directed and weighted complex networks has recently been the subject of intense study by a number of groups in wide variety of networks, worldwide. Recent studies by Sharma et al. on multi-type biological networks identified control targets in phenotypically characterized Osteosarcoma showing important role of genes and proteins responsible for maintaining tumor microenvironment.
The host-pathogen interaction is defined as how microbes or viruses sustain themselves within host organisms on a molecular, cellular, organismal or population level. This term is most commonly used to refer to disease-causing microorganisms although they may not cause illness in all hosts. Because of this, the definition has been expanded to how known pathogens survive within their host, whether they cause disease or not.

In cell biology, single-cell variability occurs when individual cells in an otherwise similar population differ in shape, size, position in the cell cycle, or molecular-level characteristics. Such differences can be detected using modern single-cell analysis techniques. Investigation of variability within a population of cells contributes to understanding of developmental and pathological processes,
Network medicine is the application of network science towards identifying, preventing, and treating diseases. This field focuses on using network topology and network dynamics towards identifying diseases and developing medical drugs. Biological networks, such as protein-protein interactions and metabolic pathways, are utilized by network medicine. Disease networks, which map relationships between diseases and biological factors, also play an important role in the field. Epidemiology is extensively studied using network science as well; social networks and transportation networks are used to model the spreading of disease across populations. Network medicine is a medically focused area of systems biology.













